3. Combining BioServices with external tools¶
This section shows how to use BioServices as an intermediate tool that fetch data to be used by third-party software/application.
The external applications used in this section are not part of BioServices therefore we do not provide instructions for the installation. Reader should refer to the application web site instead (URLs are provided here below). However, we indicate the way we installed them.
3.1. PYMOL¶
This example below uses the external software called PyMOL. We have installed it without trouble by downloading the source file from their website. Then, we typed those commands in a shell:
bunzip pymol-v1.6alpha1.tar.bz2
tar xvf pymol-v1.6alpha1.tar
cd pymol
python setup.py install
You may need to install library if requested. Tested under Fedora 15.
The following code uses BioServices to get the PDB Identifier of a protein
called ZAP70. To do so, we use bioservices.uniprot.UniProt to get its accession number (P43403) and its
PDB identifer. Then, we use bioservices.pdb.PDB to get the 3D structure in PDB
format.
The script above uses PyMOL in a script manner to save the 3D graphical representation of the protein (here below) but you could also use PyMOL in an interactive mode.

3.2. BioPython¶
BioPython provides many tools for IO, algorithms and access to Web services. BioServices provides access to many web services. This example shows how (i) to use BioServices to retrieve FASTA files and (ii) BioPython to play with the sequences.
Note
We assume you have installed BioPython (pip install biopython)
First, let us retrieve two FASTA sequences and save them in 2 files:
from bioservices import UniProt
u = UniProt()
akt1 = u.retrieve("P31749", "fasta")
akt2 = u.retrieve("P31751", "fasta")
fh = open("akt1.fasta", "w")
fh.write(akt1)
fh.close()
fh = open("akt2.fasta", "w")
fh.write(akt2)
fh.close()
Now, on the BioPython side, we read the 2 sequences and introspect them:
>>> from Bio import AlignIO
>>> record1 = SeqIO.read("akt1.fasta", "fasta")
>>> record2 = SeqIO.read("akt2.fasta", "fasta")
>>> record1 += "-" # this is to have 2 sequences on same length as requested by the following function
>>> alignment = AlignIO.MultipleSeqAlignment([])
>>> alignment.append(record1)
>>> alignment.append(record2)
>>> for record in alignment:
>>> print(description)
sp|P31749|AKT1_HUMAN RAC-alpha serine/threonine-protein kinase OS=Homo sapiens GN=AKT1 PE=1 SV=2
sp|P31751|AKT2_HUMAN RAC-beta serine/threonine-protein kinase OS=Homo sapiens GN=AKT2 PE=1 SV=2
You are ready to play with BioPython multiple alignment tools. Please consult BioPython documentation for more examples.
3.3. Galaxy¶
- URL
- Date
Aug 2013
Galaxy is an open, web-based platform for accessible, reproducible, and transparent computational biomedical research. It provides worflows and plugins to many web resources.
This tutorial shows how to link bioservices and galaxy. Our tutorial will provide a plugin to Galaxy so that a user can retrieve a FASTA file via BioServices and the wrapping of UniProt Web Services.
We assume that you installed Galaxy on your system via the source code:
hg clone https://bitbucket.org/galaxy/galaxy-dist/
cd galaxy-dist
hg update stable
The tree directory should therefore contains a directory called tools/ and in the main directory, an XML file called conf_tools.py
We will first create a plugin for bioservices. This is done by adding a directory called bioservices in ./tools:
mkdir tools/bioservices
In this directory, we will create two files called uniprot.py that will contain the actual code that calls bioservices and a second XML file that will allows us to design the plugin layout.
Let us start with the plugin. It is very simple since only the UniProt Entry is required. The output will simply be the FASTA file that would have been fetched.
The XML file is:
<tool id="bioservices_uniprot" name="Get FASTA" version="1.1.0">
<description>from UniProt via Bioservices</description>
<requirements>
<requirement type="package">bioservices</requirement>
</requirements>
<command interpreter="python">uniprot.py $uniprot_id $output</command>
<inputs>
<param name="uniprot_id" type="text" label="UniProt ID" size="40" help="Provide a valid UniProt Entry (e.g. P43403) "/>
</inputs>
<outputs>
<data format="fasta" name="output" />
</outputs>
<help>
Fetch a FASTA file using UniProt via BioServices. Simply provide a valid Uniprot Entry (e.g., P43403)
</help>
</tool>
The python code will take as an input the UniProt ID and create a file that contains the FASTA data:
import sys
def __main__():
ids = sys.argv[1]
filename = sys.argv[2]
# TODO: check the validity and format ?
try:
from bioservices import UniProt
u = UniProt(verbose=False)
u.debugLevel = "ERROR"
except ImportError:
print("Could not import bioservoces ? Check that it is installed. Try 'pip install bioservices'")
try:
fasta = u.searchUniProtId(ids, "fasta")
except:
print("An error occured while fetching the FASTA file from uniprot")
try:
fh = open(filename, "w")
fh.write(fasta)
finally, you need to make Galaxy aware of this new plugin. this is done in the file called conf_tool.xml. Add bioservices plugin. The beginning of the file should look like:
<?xml version="1.0"?>
<toolbox>
<section name="Get Data" id="getext">
<tool file="bioservices/uniprot.xml"/>
<tool file="data_source/upload.xml"/>
...
Once done. start you galaxy server. The following image show the outcome: in the left hand side, you can select the bioservices plugin. Then, in the center, you can enter a uniprot entry. Press the execute button and the new file should appear in the right hand side. From there you can use Galaxy other tools to analyse the file.
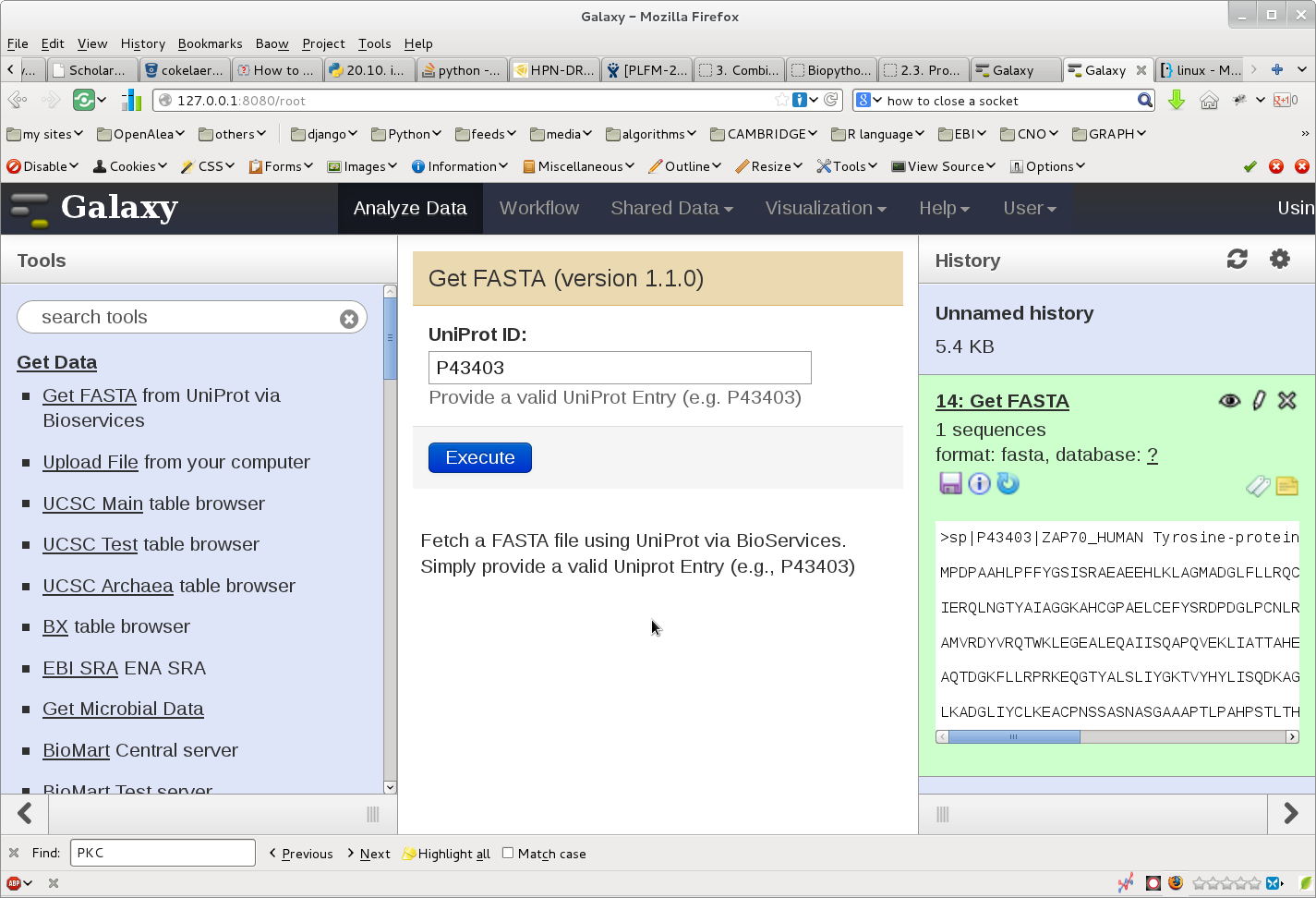
This example shows that it is possible to link Galaxy and BioServices to access to various Web Services that are available through Bioservices.