Contents
7. Utilities¶
7.1. Service module (REST or WSDL)¶
Modules with common tools to access web resources
- class REST(name, url=None, verbose=True, cache=False, requests_per_sec=3, proxies=[], cert=None, url_defined_later=False)[source]¶
The ideas (sync/async) and code using requests were inspired from the chembl python wrapper but significantly changed.
Get one value:
>>> from bioservices import REST >>> s = REST("test", "https://www.ebi.ac.uk/chemblws") >>> res = s.get_one("targets/CHEMBL2476.json", "json") >>> res['organism'] u'Homo sapiens'
The caching has two major interests. First one is that it speed up requests if you repeat requests.
>>> s = REST("test", "https://www.ebi.ac.uk/chemblws") >>> s.CACHING = True >>> # requests will be stored in a local sqlite database >>> s.get_one("targets/CHEMBL2476") >>> # Disconnect your wiki and any network connections. >>> # Without caching you cannot fetch any requests but with >>> # the CACHING on, you can retrieve previous requests: >>> s.get_one("targets/CHEMBL2476")
Advantages of requests over urllib
requests length is not limited to 2000 characters http://www.g-loaded.eu/2008/10/24/maximum-url-length/
There is no need for authentication if the web services available in bioservices except for a few exception. In such case, the username and password are to be provided with the method call. However, in the future if a services requires authentication, one can set the attribute
authenticationto a tuple:s = REST() s.authentication = ('user', 'pass')
Note about headers and content type. The Accept header is used by HTTP clients to tell the server what content types they will accept. The server will then send back a response, which will include a Content-Type header telling the client what the content type of the returned content actually is. When using the
get__headers(), you can see the User-Agent, the Accept and Content-Type keys. So, here the HTTP requests also contain Content-Type headers. In POST or PUT requests the client is actually sendingdata to the server as part of the request, and the Content-Type header tells the server what the data actually is For a POST request resulting from an HTML form submission, the Content-Type of the request should be one of the standard form content types: application/x-www-form-urlencoded (default, older, simpler) or multipart/form-data (newer, adds support for file uploads)Constructor
- Parameters
name (str) – a name for this service
url (str) – its URL
verbose (bool) – prints informative messages if True (default is True)
requests_per_sec – maximum number of requests per seconds are restricted to 3. You can change that value. If you reach the limit, an error is raise. The reason for this limitation is that some services (e.g.., NCBI) may black list you IP. If you need or can do more (e.g., ChEMBL does not seem to have restrictions), change the value. You can also have several instance but again, if you send too many requests at the same, your future requests may be retricted. Currently implemented for REST only
All instances have an attribute called
loggingthat is an instanceof theloggingmodule. It can be used to print information, warning, error messages:self.logging.info("informative message") self.logging.warning("warning message") self.logging.error("error message")
The attribute
debugLevelcan be used to set the behaviour of the logging messages. If the argument verbose is True, the debugLebel is set to INFO. If verbose if False, the debugLevel is set to WARNING. However, you can use thedebugLevelattribute to change it to one of DEBUG, INFO, WARNING, ERROR, CRITICAL. debugLevel=WARNING means that only WARNING, ERROR and CRITICAL messages are shown.- property TIMEOUT¶
- content_types = {'bed': 'text/x-bed', 'default': 'application/x-www-form-urlencoded', 'fasta': 'text/x-fasta', 'gff3': 'text/x-gff3', 'gif': 'image/gif', 'jpeg': 'image/jpg', 'jpg': 'image/jpg', 'json': 'application/json', 'jsonp': 'text/javascript', 'nh': 'text/x-nh', 'phylip': 'text/x-phyloxml+xml', 'phyloxml': 'text/x-phyloxml+xml', 'png': 'image/png', 'seqxml': 'text/x-seqxml+xml', 'svg': 'image/svg', 'svg+xml': 'image/svg+xml', 'text': 'text/plain', 'txt': 'text/plain', 'xml': 'application/xml', 'yaml': 'text/x-yaml'}¶
- get_headers(content='default')[source]¶
- Parameters
content (str) – set to default that is application/x-www-form-urlencoded so that it has the same behaviour as urllib2 (Sept 2014)
- get_one(query=None, frmt='json', params={}, **kargs)[source]¶
if query starts with http:// do not use self.url
- http_get(query, frmt='json', params={}, **kargs)[source]¶
query is the suffix that will be appended to the main url attribute.
query is either a string or a list of strings.
if list is larger than ASYNC_THRESHOLD, use asynchronous call.
- http_post(query, params=None, data=None, frmt='xml', headers=None, files=None, content=None, **kargs)[source]¶
- property session¶
- class Service(name, url=None, verbose=True, requests_per_sec=10, url_defined_later=False)[source]¶
Base class for WSDL and REST classes
See also
Constructor
- Parameters
name (str) – a name for this service
url (str) – its URL
verbose (bool) – prints informative messages if True (default is True)
requests_per_sec – maximum number of requests per seconds are restricted to 3. You can change that value. If you reach the limit, an error is raise. The reason for this limitation is that some services (e.g.., NCBI) may black list you IP. If you need or can do more (e.g., ChEMBL does not seem to have restrictions), change the value. You can also have several instance but again, if you send too many requests at the same, your future requests may be retricted. Currently implemented for REST only
All instances have an attribute called
loggingthat is an instanceof theloggingmodule. It can be used to print information, warning, error messages:self.logging.info("informative message") self.logging.warning("warning message") self.logging.error("error message")
The attribute
debugLevelcan be used to set the behaviour of the logging messages. If the argument verbose is True, the debugLebel is set to INFO. If verbose if False, the debugLevel is set to WARNING. However, you can use thedebugLevelattribute to change it to one of DEBUG, INFO, WARNING, ERROR, CRITICAL. debugLevel=WARNING means that only WARNING, ERROR and CRITICAL messages are shown.- property CACHING¶
- easyXML(res)[source]¶
- Use this method to convert a XML document into an
easyXMLobject
The easyXML object provides utilities to ease access to the XML tag/attributes.
Here is a simple example starting from the following XML
>>> from bioservices import * >>> doc = "<xml> <id>1</id> <id>2</id> </xml>" >>> s = Service("name") >>> res = s.easyXML(doc) >>> res.findAll("id") [<id>1</id>, <id>2</id>]
- property easyXMLConversion¶
If True, xml output from a request are converted to easyXML object (Default behaviour).
- pubmed(Id)[source]¶
Open a pubmed Id into a browser tab
- Parameters
Id – a valid pubmed Id in string or integer format.
The URL is a concatenation of the pubmed URL http://www.ncbi.nlm.nih.gov/pubmed/ and the provided Id.
- response_codes = {200: 'OK', 201: 'Created', 400: 'Bad Request. There is a problem with your input', 404: 'Not found. The resource you requests does not exist', 405: 'Method not allowed', 406: 'Not Acceptable. Usually headers issue', 410: 'Gone. The resource you requested was removed.', 415: 'Unsupported Media Type', 500: 'Internal server error. Most likely a temporary problem', 503: 'Service not available. The server is being updated, try again later'}¶
some useful response codes
- property url¶
URL of this service
- class WSDLService(name, url, verbose=True, cache=False)[source]¶
Class dedicated to the web services based on WSDL/SOAP protocol.
See also
RESTService,ServiceConstructor
- Parameters
The
servgive access to all WSDL functionalities of the service.The
methodsis an alias to self.serv.methods and returns the list of functionalities.- property TIMEOUT¶
- property wsdl_methods¶
returns methods available in the WSDL service
7.2. xmltools module¶
This module includes common tools to manipulate XML files
- class easyXML(data, encoding='utf-8')[source]¶
class to ease the introspection of XML documents.
This class uses the standard xml module as well as the package BeautifulSoup to help introspecting the XML documents.
>>> from bioservices import * >>> n = ncbiblast.NCBIblast() >>> res = n.getParameters() # res is an instance of easyXML >>> # You can retreive XML from this instance of easyXML and print the content >>> # in a more human-readable way. >>> res.soup.findAll('id') # a Beautifulsoup instance is available >>> res.root # and the root using xml.etree.ElementTree
There is a getitem so you can type:
res['id']
which is equivalent to:
res.soup.findAll('id')
There is also aliases findAll and prettify.
Constructor
- Parameters
data – an XML document format
fixing_unicode – use only with HGNC service to fix issue with the XML returned by that particular service. No need to use otherwise. See
HGNCdocumentation for details.encoding – default is utf-8 used. Used to fix the HGNC XML only.
The data parameter must be a string containing the XML document. If you have an URL instead, use
readXML- getchildren()[source]¶
returns all children of the root XML document
This is just an alias to self.soup.getchildren()
- property soup¶
Returns the beautiful soup instance
- class readXML(url, encoding='utf-8')[source]¶
Read XML and converts to beautifulsoup data structure
easyXML accepts as input a string. This class accepts a filename instead inherits from easyXML
See also
Constructor
- Parameters
data – an XML document format
fixing_unicode – use only with HGNC service to fix issue with the XML returned by that particular service. No need to use otherwise. See
HGNCdocumentation for details.encoding – default is utf-8 used. Used to fix the HGNC XML only.
The data parameter must be a string containing the XML document. If you have an URL instead, use
readXML
8. Services¶
8.1. ArrayExpress¶
8.2. Biocontainers¶
Interface to biocontainer
What is biocontainers
- URL
- Citation
BioContainers is an open-source project that aims to create, store, and distribute bioinformatics software containers and packages.
—From biocontainers (about), Jan 2021
8.3. BiGG¶
Interface to the BiGG Models API Service
What is BiGG Models?
“BiGG Models is a knowledgebase of genome-scale metabolic network reconstructions. BiGG Models integrates more than 70 published genome-scale metabolic networks into a single database with a set of standardized identifiers called BiGG IDs. Genes in the BiGG models are mapped to NCBI genome annotations, and metabolites are linked to many external databases (KEGG, PubChem, and many more).”
—BiGG Models Home Page, March 10, 2020.
- class BiGG(verbose=False, cache=False)[source]¶
Interface to the BiGG Models <http://bigg.ucsd.edu/> API Service.
>>> from bioservices import BiGG >>> bigg = BiGG() >>> bigg.search("e coli", "models") [{'bigg_id': 'e_coli_core', 'gene_count': 137, 'reaction_count': 95, 'organism': 'Escherichia coli str. K-12 substr. MG1655', 'metabolite_count': 72}, ... ]
- property models¶
- property version¶
8.4. BioDBnet¶
This module provides a class BioDBNet to access to BioDBNet WS.
What is BioDBNet ?
- URL
- Service
- Citations
Mudunuri,U., Che,A., Yi,M. and Stephens,R.M. (2009) bioDBnet: the biological database network. Bioinformatics, 25, 555-556
“BioDBNet Database is a repository hosting computational models of biological systems. A large number of the provided models are published in the peer-reviewed literature and manually curated. This resource allows biologists to store, search and retrieve mathematical models. In addition, those models can be used to generate sub-models, can be simulated online, and can be converted between different representational formats. “
—From BioDBNet website, Dec. 2012
New in version 1.2.3.
Section author: Thomas Cokelaer, Feb 2014
- class BioDBNet(verbose=True, cache=False)[source]¶
Interface to the BioDBNet service
>>> from bioservices import * >>> s = BioDBNet()
Most of the BioDBNet WSDL are available. There are functions added to the original interface such as
extra_getReactomeIds().Use
db2db()to convert from 1 database to some databases. UsedbReport()to get the convertion from one database to all databases.Constructor
- Parameters
verbose (bool) –
- db2db(input_db, output_db, input_values, taxon=9606)[source]¶
Retrieves models associated to the provided Taxonomy text.
- Parameters
input_db – input database.
output_db – list of databases to map to.
input_values – list of identifiers to map to the output databases
- Returns
dataframe where index correspond to the input database identifiers. The columns contains the identifiers for each output database (see example here below).
>>> from bioservices import BioDBNet >>> input_db = 'Ensembl Gene ID' >>> output_db = ['Gene Symbol'] >>> input_values = ['ENSG00000121410', 'ENSG00000171428'] >>> df = s.db2db(input_db, output_db, input_values, 9606) Gene Symbol Ensembl Gene ID ENSG00000121410 A1BG ENSG00000171428 NAT1
- dbFind(output_db, input_values, taxon='9606')[source]¶
dbFind method
dbFind can be used when you do not know the actual type of your identifiers or when you have a mixture of different types of identifiers. The tool finds the identifier type and converts them into the selected output if the identifiers are within the network.
- Parameters
- Returns
a dataframe with index set to the input values.
>>> b.dbFind("Gene ID", ["ZMYM6_HUMAN", "NP_710159", "ENSP00000305919"]) Gene ID Input Type InputValue ZMYM6_HUMAN 9204 UniProt Entry Name NP_710159 203100 RefSeq Protein Accession ENSP00000305919 203100 Ensembl Protein ID
- dbOrtho(input_db, output_db, input_values, input_taxon, output_taxon)[source]¶
Convert identifiers from one species to identifiers of a different species
- Parameters
input_db – input database
output_db – output database
input_values – list of identifiers to retrieve
input_taxon – input taxon
output_taxon – output taxon
- Returns
dataframe where index correspond to the input database identifiers. The columns contains the identifiers for each output database (see example here below)
>>> df = b.dbOrtho("Gene Symbol", "Gene ID", ["MYC", "MTOR", "A1BG"], ... input_taxon=9606, output_taxon=10090) Gene ID InputValue 0 17869 MYC 1 56717 MTOR 2 117586 A1BG
- dbReport(input_db, input_values, taxon=9606)[source]¶
Same as
db2db()but returns results for all possible outputs.- Parameters
input_db – input database
input_values – list of identifiers to retrieve
- Returns
dataframe where index correspond to the input database identifiers. The columns contains the identifiers for each output database (see example here below)
df = s.dbReport("Ensembl Gene ID", ['ENSG00000121410', 'ENSG00000171428'])
- dbWalk(db_path, input_values, taxon=9606)[source]¶
Walk through biological database network
dbWalk is a form of database to database conversion where the user has complete control on the path to follow while doing the conversion. When a input/node is added to the path the input selection gets updated with all the nodes that it can access directly.
- Parameters
db_path – path to follow in the databases
input_values – list of identifiers
- Returns
a dataframe with columns corresponding to the path nodes
A typical example is to get the Ensembl mouse homologs for Ensembl Gene ID’s from human. This conversion is not possible through
db2db()as Homologene does not have Ensembl ID’s and the input and output nodes to acheive this would both be ‘Ensembl Gene ID’. It can however be run by using dbWalk as follows. Add Ensembl Gene ID to the path, then add Gene Id, Homolog - Mouse Gene ID and Ensebml Gene ID to complete the path.db_path = "Ensembl Gene ID->Gene ID->Homolog - Mouse Gene ID->Ensembl Gene ID" s.dbWalk(db_path, ["ENSG00000175899"])
Todo
check validity of the path
- getDirectOutputsForInput(input_db)[source]¶
Gets all the direct output nodes for a given input node
Gets all the direct output nodes for a given input node Outputs reachable by single edge connection in the bioDBnet graph.
b.getDirectOutputsForInput("genesymbol") b.getDirectOutputsForInput("Gene Symbol") b.getDirectOutputsForInput("pdbid") b.getDirectOutputsForInput("PDB ID")
8.5. BioGrid¶
This module provides a class BioGrid.
What is BioGrid ?
- URL
- Service
Via the PSICQUIC class
BioGRID is an online interaction repository with data compiled through comprehensive curation efforts. Our current index is version 3.2.97 and searches 37,954 publications for 638,453 raw protein and genetic interactions from major model organism species. All interaction data are freely provided through our search index and available via download in a wide variety of standardized formats.
—From BioGrid website, Feb. 2013
- class BioGRID(query=None, taxId=None, exP=None)[source]¶
Interface to BioGRID.
>>> from bioservices import BioGRID >>> b = BioGRID(query=["map2k4","akt1"],taxId = "9606") >>> interactors = b.biogrid.interactors
Examples:
>>> from bioservices import BioGRID >>> b = BioGRID(query=["mtor","akt1"],taxId="9606",exP="two hybrid") >>> b.biogrid.interactors
One can also query an entire organism, by using the taxid as the query:
>>> b = BioGRID(query="6239")
8.6. BioMart¶
This module provides a class BioModels that allows an easy access
to all the BioModel service.
What is BioMart ?
The BioMart project provides free software and data services to the international scientific community in order to foster scientific collaboration and facilitate the scientific discovery process. The project adheres to the open source philosophy that promotes collaboration and code reuse.
—from BioMart March 2013
Note
SOAP and REST are available. We use REST for the wrapping.
- class BioMart(host=None, verbose=False, cache=False, secure=False)[source]¶
Interface to the BioMart service
BioMart is made of different views. Each view correspond to a specific MART. For instance the UniProt service has a BioMart view.
The registry can help to find the different services available through BioMart.
>>> from bioservices import * >>> s = BioMart() >>> ret = s.registry() # to get information about existing services
The registry is a list of dictionaries. Some aliases are available to get all the names or databases:
>>> s.names # alias to list of valid service names from registry >>> "unimart" in s.names True
Once you selected a view, you will want to select a database associated with this view and then a dataset. The datasets can be retrieved as follows:
>>> s.datasets("prod-intermart_1") # retrieve datasets available for this mart
The main issue is how to figure out the database name (here prod-intermart_1) ? Indeed, from the web site, what you see is the displayName and you must introspect the registry to get this information. In BioServices, we provide the
lookfor()method to help you. For instance, to retrieve the database name of interpro, type:>>> s = BioMart(verbose=False) >>> s.lookfor("interpro") Candidate: database: intermart_1 MART name: prod-intermart_1 displayName: INTERPRO (EBI UK) hosts: www.ebi.ac.uk
The display name (INTERPRO) correspond to the MART name prod-intermart_1. Let us you it to retrieve the datasets:
>>> s.datasets("prod-intermart_1") ['protein', 'entry', 'uniparc']
Now that we have the dataset names, we can select one and build a query. Queries are XML that contains the dataset name, some attributes and filters. The dataset name is one of the element returned by the datasets method. Let us suppose that we want to query protein, we need to add this dataset to the query:
>>> s.add_dataset_to_xml("protein")
Then, you can add attributes (one of the keys of the dictionary returned by attributes(“protein”):
>>> s.add_attribute_to_xml("protein_accession")
Optional filters can be used:
>>> s.add_filter_to_xml("protein_length_greater_than", 1000)
Finally, you can retrieve the XML query:
>>> xml_query = s.get_xml()
and send the request to biomart:
>>> res = s.query(xml_query) >>> len(res) 12801 # print the first 10 accession numbers >>> res = res.split("\n") >>> for x in res[0:10]: print(x) ['P18656', 'Q81998', 'O09585', 'O77624', 'Q9R3A1', 'E7QZH5', 'O46454', 'Q9T3F4', 'Q9TCA3', 'P72759']
REACTOME example:
s.lookfor("reactome") s.datasets("REACTOME") ['interaction', 'complex', 'reaction', 'pathway'] s.new_query() s.add_dataset_to_xml("pathway") s.add_filter_to_xml("species_selection", "Homo sapiens") s.add_attribute_to_xml("pathway_db_id") s.add_attribute_to_xml("_displayname") xmlq = s.biomartQuery.get_xml() res = s.query(xmlq)
Note
the biomart sevice is slow (in my experience, 2013-2014) so please be patient…
Constructor
URL required to use biomart change quite often. Experience has shown that BioMart class in Bioservices may fail. This is not a bioservices issue but due to API changes on server side.
For that reason the host is not filled anymore and one must set it manually.
Let us take the example of the ensembl biomart. The host is
www.ensembl.org
Note that there is no prefix http and that the actual URL looked for internally is http://www.ensembl.org/biomart/martview
(It used to be martservice in 2012-2016)
Another reason to not set any default host is that servers may be busy or take lots of time to initialise (if many MARTS are available). Usually, one knows which MART to look at, in which case you may want to use a specific host (e.g., www.ensembl.org) that will speed up significantly the initialisation time.
- Parameters
host (str) – a valid host (e.g. “www.ensembl.org”, gramene.org)
List of databases are available in this webpage http://www.biomart.org/community.html
- attributes(dataset)[source]¶
to retrieve attributes available for a dataset:
- Parameters
dataset (str) – e.g. oanatinus_gene_ensembl
- configuration(dataset)[source]¶
to retrieve configuration available for a dataset:
- Parameters
dataset (str) – e.g. oanatinus_gene_ensembl
- property databases¶
list of valid datasets
- datasets(mart, raw=False)[source]¶
to retrieve datasets available for a mart:
- Parameters
mart (str) – e.g. ensembl. see
namesfor a list of valid MART names the mart is the database. see lookfor method or databases attributes
>>> s = BioMart(verbose=False) >>> s.datasets("prod-intermart_1") ['protein', 'entry', 'uniparc']
- property displayNames¶
list of valid datasets
- filters(dataset)[source]¶
to retrieve filters available for a dataset:
- Parameters
dataset (str) – e.g. oanatinus_gene_ensembl
>>> s.filters("uniprot").split("\n")[1].split("\t") >>> s.filters("pathway")["species_selection"] [Arabidopsis thaliana,Bos taurus,Caenorhabditis elegans,Canis familiaris,Danio rerio,Dictyostelium discoideum,Drosophila melanogaster,Escherichia coli,Gallus gallus,Homo sapiens,Mus musculus,Mycobacterium tuberculosis,Oryza sativa,Plasmodium falciparum,Rattus norvegicus,Saccharomyces cerevisiae,Schizosaccharomyces pombe,Staphylococcus aureus N315,Sus scrofa,Taeniopygia guttata ,Xenopus tropicalis]
- property host¶
- property hosts¶
list of valid hosts
- property marts¶
list of marts
- property names¶
list of valid datasets
- query(xmlq)[source]¶
Send a query to biomart
The query must be formatted in a XML format which looks like ( example from https://gist.github.com/keithshep/7776579):
<?xml version="1.0" encoding="UTF-8"?> <!DOCTYPE Query> <Query virtualSchemaName="default" formatter="CSV" header="0" uniqueRows="0" count="" datasetConfigVersion="0.6"> <Dataset name="mmusculus_gene_ensembl" interface="default"> <Filter name="ensembl_gene_id" value="ENSMUSG00000086981"/> <Attribute name="ensembl_gene_id"/> <Attribute name="ensembl_transcript_id"/> <Attribute name="transcript_start"/> <Attribute name="transcript_end"/> <Attribute name="exon_chrom_start"/> <Attribute name="exon_chrom_end"/> </Dataset> </Query>Warning
the input XML must be valid. THere is no validation made in thiss method.
- registry()[source]¶
to retrieve registry information
the XML contains list of children called MartURLLocation made of attributes. We parse the xml to return a list of dictionary. each dictionary correspond to one MART.
aliases to some keys are provided: names, databases, displayNames
- property valid_attributes¶
list of valid datasets
8.7. BioModels¶
This module provides a class BioModels to access to BioModels WS.
What is BioModels ?
- URL
- Service
- Citations
please visit https://www.ebi.ac.uk/biomodels/citation for details
“BioModels is a repository of mathematical models of biological and biomedical systems. It hosts a vast selection of existing literature-based physiologically and pharmaceutically relevant mechanistic models in standard formats. Our mission is to provide the systems modelling community with reproducible, high-quality, freely-accessible models published in the scientific literature.”
—From BioModels website, March 2020
- class BioModels(verbose=True)[source]¶
Interface to the BioModels service
from bioservices import BioModels bm = BioModels() model = bm.get_model('BIOMD0000000299')
Previous API had several functions such as getAuthorsByModelId. This is easy to mimic with the new API:
bm = BioModels() models = bm.get_all_models() [x['submitter'] for x in res if x[] == "MODEL1204280003"][0]
This is also true for getDateLastModifByModelId and getModelNameById if one use the field lastModified or name. There was the ability to search for models based on their CHEBI identifiers, which is not supported anymore; this concerns functions getModelsIdByChEBI, getModelsIdByChEBIId, getSimpleModelsByChEBIIds, getSimpleModelsRelatedWithChEBI. For other searches related to Reactome, Uniprot identifiers or GO terms, the
search()method should work:bm.search("P10113") bm.search("REACT_33") bm.search("GO:0006919")
constructor
- Parameters
verbose (bool) –
- get_model(model_id, frmt='json')[source]¶
Fetch information about a given model at a particular revision.
- get_model_download(model_id, filename=None, output_filename=None)[source]¶
Download a particular file associated with a given model or all its files as a COMBINE archive.
- Parameters
- Returns
nothing. This function save the model into a ZIP file called after the model identifier. If parameter filename is specified, then the output file is the requested filename (if found)
bm.get_model_download("BIOMD0000000100", filename="BIOMD0000000100.png") bm.get_model_download("BIOMD0000000100")
This function can retrieve all files in a ZIP archive or a single image. In the example below, we retrieve the PNG and plot it using matplotlib. Using your favorite image viewver, you should get a better resolution. Or just download the SVG version of the model.
from bioservices import BioModels bm = BioModels() from easydev import TempFile with TempFile(suffix=".png") as fout: bm.get_model_download("BIOMD0000000100", filename="BIOMD0000000100.png", output_filename=fout.name) from pylab import imshow, imread imshow(imread(fout.name), aspect="auto")
(Source code, png, hires.png, pdf)
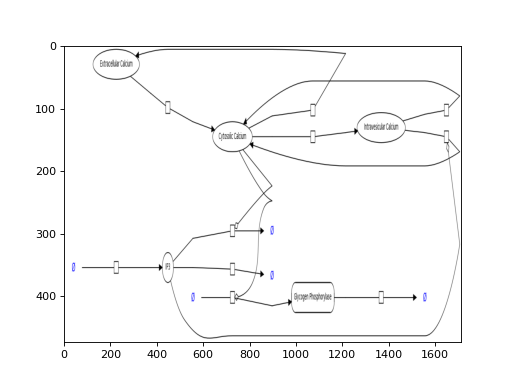
- get_model_files(model_id, frmt='json')[source]¶
Extract metadata information of model files of a particular model
- Parameters
model_id – a valid BioModels identifier
frmt – format of the output (json, xml)
- get_p2m_missing(frmt='json')[source]¶
Retrieve all models in Path2Models that are now only available indirectly, through the representative model for the corresponding genus
- Parameters
frmt (str) – the format of the result (xml, csv, json)
- Returns
list of model identifiers
- get_p2m_representative(model, frmt='json')[source]¶
Retrieve a representative model in Path2Models
Get the representative model identifier for a given missing model in Path2Models. This endpoint accepts as parameters a mandatory model identifier and an optional response format
- get_p2m_representatives(models, frmt='json')[source]¶
Find the replacement accessions for a set of Path2Models entries
Get the representative model identifiers of a set of given missing models in Path2Models. This end point expects a comma-separated list of model identifiers (without any surrounding whitespace) and an optional response format. Examples: BMID000000112902, BMID000000009880, BMID000000027397.
- Parameters
from bioservices import BioModels bm = BioModels() bm.get_p2m_representatives("BMID000000112902, BMID000000009880, BMID000000027397")
- get_pdgsmm_missing(frmt='json')[source]¶
Retrieve the identifiers of all PDGSMM entries that are no longer directly accessible
- Parameters
frmt (str) – the format of the result (xml, csv, json)
- Returns
list of model identifiers
- get_pdgsmm_representative(model, frmt='json')[source]¶
Retrieve a representative model in PDGSMM
Get the representative model identifier for a given missing model in PDGSMM. This endpoint accepts as parameters a mandatory model identifier and an optional response format.
- get_pdgsmm_representatives(models, frmt='json')[source]¶
Find the replacement accessions for a set of PDFSSM
Get the representative model identifiers of a set of given missing models in PDGSMM. This end point expects a comma-separated list of model identifiers (without any surrounding whitespace) and an optional response format. Examples: MODEL1707110145,MODEL1707112456,MODEL1707115900.
- search(query, offset=None, numResults=None, sort=None, frmt='json')[source]¶
Search models of interest via keywords.
Examples: PUBMED:”27869123” to search models associated with the PubMed record identified by 27869123.
- Parameters
query (str) – search query. colon character must be escaped
offset (int) – number of items to skip before starting to collect the result set
numResults (int) – number of items to return
sort (str) – sort criteria in {id-asc, relevance-asc, relevance-desc, first_author-asc, first_author, name-asc, name-desc, publication_year-asc, publication_year-desc}
frmt (str) – format of the output (json, xml)
- search_download(models, output_filename='models.zip', force=False)[source]¶
Returns models (XML) corresponding to a list of model identifiers.
- Parameters
Todo
if no models are found (typos), an error message is printed. if one model is not found, there is no warning or errors. Could be nice to have a warning by introspecting the number of models in the output file
- search_parameter(query, start=0, size=10, sort=None, frmt='json')[source]¶
Search for parameters of a model
Details BioModels Parameters is a resource that facilitates easy search and retrieval of parameter values used in the SBML models stored in the BioModels repository. Users can search for a model entity (e.g. a protein or drug) to retrieve the rate equations describing it; the associated parameter values and the initial concentration from the SBML models in BioModels. Although these data are directly extracted from the curated SBML models, they are not individually curated or validated; rather presented as such in the table below. Hence BioModels Parameters table will only provide a quick overview of available parameter values for guidance and original model should be referred to understand the complete context of the parameter usage.
- Parameters
bm.search_parameter("MAPK", size=100, sort="entity")
8.8. ChEBI¶
This module provides a class ChEBI
What is ChEBI
“The database and ontology of Chemical Entities of Biological Interest
—From ChEBI web page June 2013
- class ChEBI(verbose=False)[source]¶
Interface to ChEBI
>>> from bioservices import * >>> ch = ChEBI() >>> res = ch.getCompleteEntity("CHEBI:27732") >>> res.smiles CN1C(=O)N(C)c2ncn(C)c2C1=O
Constructor
- Parameters
verbose (bool) –
- conv(chebiId, target)[source]¶
Calls
getCompleteEntity()and returns the identifier of a given database- Parameters
chebiId (str) – a valid ChEBI identifier (string)
target – the identifier of the database
- Returns
the identifier
>>> ch.conv("CHEBI:10102", "KEGG COMPOUND accession") ['C07484']
- getAllOntologyChildrenInPath(chebiId, relationshipType, onlyWithChemicalStructure=False)[source]¶
Retrieves the ontology children of an entity including the relationship type
- Parameters
>>> ch.getAllOntologyChildrenInPath("CHEBI:27732", "has part")
- getCompleteEntity(chebiId)[source]¶
Retrieves the complete entity including synonyms, database links and chemical structures, using the ChEBI identifier.
- param str chebiId
a valid ChEBI identifier (string)
- return
an object containing fields such as mass, names, smiles
>>> from bioservices import * >>> ch = ChEBI() >>> res = ch.getCompleteEntity("CHEBI:27732") >>> res.mass 194.19076
The returned structure is the raw object returned by the API. You can extract names from other sources for instance:
>>> [x[0] for x in res.DatabaseLinks if x[1].startswith("KEGG")] [C07481, D00528] >>> [x[0] for x in res.DatabaseLinks if x[1].startswith("ChEMBL")] [116485]
See also
- getCompleteEntityByList(chebiIdList=[])[source]¶
Given a list of ChEBI accession numbers, retrieve the complete Entities.
The maximum size of this list is 50.
See also
- getLiteEntity(search, searchCategory='ALL', maximumResults=200, stars='ALL')[source]¶
Retrieves list of entities containing the ChEBI ASCII name or identifier
- Parameters
The input parameters are a search string and a search category. If the search category is null then it will search under all fields. The search string accepts the wildcard character “*” and also unicode characters. You can get maximum results upto 5000 entries at a time.
>>> ch.getLiteEntity("CHEBI:27732") [(LiteEntity){ chebiId = "CHEBI:27732" chebiAsciiName = "caffeine" searchScore = 4.77 entityStar = 3 }] >>> res = ch.getLiteEntity("caffeine") >>> res = ch.getLiteEntity("caffeine", maximumResults=10) >>> len(res) 10
See also
- getOntologyChildren(chebiId)[source]¶
Retrieves the ontology children of an entity including the relationship type
- Parameters
chebiId (str) – a valid ChEBI identifier (string)
- getOntologyParents(chebiId)[source]¶
Retrieves the ontology parents of an entity including the relationship type
- Parameters
chebiId (str) – a valid ChEBI identifier (string)
- getStructureSearch(structure, mode='MOLFILE', structureSearchCategory='SIMILARITY', totalResults=50, tanimotoCutoff=0.25)[source]¶
Does a substructure, similarity or identity search using a structure.
- Parameters
structure (str) – the input structure
mode (str) – type of input (MOLFILE, SMILES, CML” (note that the API uses type but this is a python keyword)
structureSearchCategory (str) – category of the search. Can be “SIMILARITY”, “SUBSTRUCTURE”, “IDENTITY”
totalResults (int) – limit the number of results to 50 (default)
tanimotoCuoff – limit results to scores higher than this parameter
>>> ch = ChEBI() >>> smiles = ch.getCompleteEntity("CHEBI:27732").smiles >>> ch.getStructureSearch(smiles, "SMILES", "SIMILARITY", 3, 0.25)
- getUpdatedPolymer(chebiId)[source]¶
Returns the UpdatedPolymer object
- Parameters
chebiId (str) –
chebiId – a valid ChEBI identifier (string)
- Returns
an object with information as described below.
The object contains the updated 2D MolFile structure, GlobalFormula string containing the formulae for each repeating-unit, the GlobalCharge string containing the charge on individual repeating-units and the primary ChEBI ID of the polymer, even if the secondary Identifier was passed to the web-service.
8.9. ChEMBL¶
This module provides a class ChEMBL
What is ChEMBL
“Using the ChEMBL web service API users can retrieve data from the ChEMBL database in a programmatic fashion. The following list defines the currently supported functionality and defines the expected inputs and outputs of each method.”
—From ChEMBL web page Dec 2012
- class ChEMBL(verbose=False, cache=False)[source]¶
New ChEMBL API bioservices 1.6.0
Resources
ChEMBL database is made of a set of resources. We recommend to look at https://arxiv.org/pdf/1607.00378.pdf
Here we first create an instance and retrieve the first 1000 molecules from the database using the limit parameter.
>>> from bioservices import ChEMBL >>> c = ChEMBL() >>> res = c.get_molecule(limit=1000)
The returned objet is a list of 1000 records, each of them being a dictionary. The molecule resource is actually a very large one and one may want to skip some entries. This is possible using the offset parameter as follows:
# Retrieve 1000 molecules skipping the first 50 res = c.get_molecule(limit=1000, offset=50)
If you want to know all resources available and the number of entries in each resources, use:
status = c.get_status_resources()
For instance, you should be able to get the total number of entries in the mechanism resource is about 5,000:
print(status['mechanism'])
To retrieve all entries from the mechanism resource, you can either set limit to a value large enough:
res = c.get_mechanism(limit=1000000)
or simply set it to -1:
res = c.get_mechanism(limit=-1)
All resources methods behaves in the same way.
Those resources methods are:
get_activity(),get_assay(),get_atc_class(),get_binding_site(),get_biotherapeutic(),get_cell_line(),get_chembl_id_lookup(),get_compound_record(),get_compound_structural_alert(),get_document(),get_document_similarity(),get_document_term(),get_drug(),get_drug_indication(),get_go_slim(),get_mechanism(),get_metabolism(),get_molecule(),get_molecule_form(),get_protein_class(),get_source(),get_target(),get_target_component(),get_target_prediction(),get_target_relation(),get_tissue().3 ways of getting items
Retrieve everything:
c.get_molecule(limit=-1)
Retrieve a specific entry:
c.get_molecule("CHEMBL24")
Retrieve a set of entries:
c.get_molecule(["CHEMBL24","CHEMBL2"])
Filtering and Ordering
For ordering the results, we provide a simple method
order_by()that allows to sort the dictionary according to values in a specific key.Any data returned by a resource method (a list of dictionary) can be process through this method:
c = ChEMBL() data = c.get_drug(limit=100) ordered_data = c.order_by(data, 'chirality')
If you want to order using a key within a key, for instance order by molecular weight stored in the molecular_properties key, use the double underscore method as follows:
c = ChEMBL() data = c.get_drug(limit=100) ordered_data = c.order_by(data, 'molecular_properties__mw_freebase')
For filtering, it is possible to apply search filters to any resources. For example, it is possible to return all ChEMBL targets that contain the term ‘kinase’ in the pref_name attribute:
c.get_target(filters='pref_name__contains=kinase")
The pattern for applying a filter is as follows:
[field]__[filter_type]=[value]
where field has to be found by the user. Simply introspect the content of an item returned by the resource. For instance:
c.get_target(limit=1) # to get one entry
Let us consider the case of the molecule resource. You can retrieve the first 10 molecules using e.g.:
res = c.get_molecule(limit=10)
If you look at the first entry using res[0], you will get about 38 keys. For instance molecule_properties or molecule_chembl_id.
You can filter the molecules to keep only the molecule_chembl_id that match either CHEMBL25 or CHEMBL1000 using:
res = c.get_molecule(filters='molecule_chembl_id__in=CHEMBL25,CHEMBL1000')
For molecule_properties, this is actually a dictionary. For instance, inside the molecule_properties field, you have the molecular weight (mw_freebase). So to apply this filter, you need to use the following code (to keep molecules with molecular weight greater than 300:
res = c.get_molecule(filters='molecule_properties__mw_freebase__gte=300')
Here are the different types of filtering:
Filter Type
Description
exact (iexact)
Exact match with query
contains
wild card search with query
startswith
starts with query
endswith
ends with query
regex
regulqr expression query
gt (gte)
Greater than (or equal)
lt (lte)
Less than (or equal)
range
Within a range of values
in
Appears within list of query values
isnull
Field is null
search
Special type of filter allowing a full text search based on Solr queries.
Several filters can be applied at the same time using a list:
filters = ['molecule_properties__mw_freebase__gte=300'] filters += ['molecule_properties__alogp__gte=3'] res = c.get_molecule(filters)
Use Cases: (inspired from ChEMBL documentation)
Search molecules by synonym:
>>> from bioservices import ChEMBL >>> c = ChEMBL() >>> res = c.search_molecule('aspirin')
or SMILE, or InChiKey, or CHEMBLID:
>>> res = c.get_molecule("CC(=O)Oc1ccccc1C(=O)O") >>> res = c.get_molecule("BSYNRYMUTXBXSQ-UHFFFAOYSA-N") >>> res = c.get_molecule('CHEMBL25')
Several molecules at the same time can also be retrieved using lists:
>>> res = c.get_molecule(['CHEMBL25', 'CHEMBL2'])
Search target by gene name:
>>> res = c.search_target("GABRB2") >>> len(res['targets']) 18
or directly in the target synonym field:
>>> res = c.get_target(filters='target_synonym__icontains=GABRB2')
Note
Not sure what is the difference between icontains vs contains. It looks like icontains is more permissive (you get more entries with icontains).
Having a list of molecules ChEMBL IDs in a list, get uniprot accession numbers that map to those compounds:
# First, get some IDs of approved drugs (about 2000 molecules) c = ChEMBL() drugs = c.get_approved_drugs() IDs = [x['molecule_chembl_id'] for x in drugs] # we jump from compounds to targets through activities # Here this is a one to many mapping so we initialise a default # dictionary. compound2target = defaultdict(set) filter = "molecule_chembl_id__in={}" for i in range(0, len(IDs), 50): activities = c.get_activity(filter.format(IDs[i:i+50])) # get target ChEMBL IDs from activities for act in activities: compound2target[act['molecule_chembl_id']].add(act['target_chembl_id']) # What we need is to get targets for all targets found in the previous # step. For each compound/drug there are hundreds of targets though. And # we will call the get_target for each list of hundreds targets. This # will take forever. Instead, because there are *only* 12,000 targets, # let us download all of them ! This took about 4 minutes on this test but # if you use the cache, next time it will be much much quicker. This is # not down at the activities level because there are too many entries targets = c.get_target(limit=-1) # identifies all target chembl id to easily retrieve the entry later on target_names = [target['target_chembl_id'] for target in targets] # retrieve all uniprot accessions for all targets of each compound for compound, targs in compounds2targets.items(): accessions = set() for target in targs: index = target_names.index(target) accessions = accessions.union([comp['accession'] for comp in targets[index]['target_components']]) compounds2targets[compound] = accessions
In version 1.6.0 of bioservices, you can simply use:
res = c.compounds2targets(IDs)
Get Target type count for all targets:
import collections collections.Counter([x['target_type'] for x in targets]
Find compounds similar to given SMILES query with similarity threshold of 85%:
>>> SMILE = "CN(CCCN)c1cccc2ccccc12" >>> c.get_similarity(SMILE, similarity=70)
Find compounds similar to aspirin (CHEMBL25) with similarity threshold of 70%:
# search for aspirin in all molecules and from first hist # get the ChEMBL ID >>> molecules = c.search_molecule("aspirin")['molecules'] >>> chembl_id = molecules[0]['molecule_chembl_id'] # now use the :meth:`get_similarity` given the ID >>> res = c.get_similarity(chembl_id, similarity=70)
Perform substructure search using SMILES or ChEMBID:
>>> res = c.get_substructure("CN(CCCN)c1cccc2ccccc12") >>> res = c.get_substructure("CHEMBL25")
Obtain he pChEMBL value for compound:
res = c.get_activity(filters=['pchembl_value__isnull=False', 'molecule_chembl_id=CHEMBL25'])
Obtain he pChEMBL value for compound and target:
res = c.get_activity(filters=['pchembl_value__isnull=False', 'molecule_chembl_id=CHEMBL25', 'target_chembl_id=CHEMBL612545'])
Get all approved drugs:
c.get_approved_drugs(max_phase=4)
Get approved drugs for lung cancer
The ChEMBL API significantly changed in 2018 and the nez version of bioservices (1.6.0) had to change the API as well, which has been simplified.
Here below are some correspondances between the previous and the new API.
bioservices before 1.6.0
After 1.6.0
get_compounds_substructure
get_substructure
get_compounds_similar_to_SMILES
get_similarity(SMILE)
get_compounds_by_chemblId(ID)
get_similarity(ID)
get_individual_compounds_by_inChiKey
get_molecule(inchikey)
get_compounds_by_chemblId_form
get_molecule_form
get_compounds_by_chemblId_drug_mechanism
get_mechanism(ID)
get_target_by_chemblId(ID)
get_target(ID)
get_image_of_compounds_by_chemblId
get_image
etc
Constructor
- Parameters
name (str) – a name for this service
url (str) – its URL
verbose (bool) – prints informative messages if True (default is True)
requests_per_sec – maximum number of requests per seconds are restricted to 3. You can change that value. If you reach the limit, an error is raise. The reason for this limitation is that some services (e.g.., NCBI) may black list you IP. If you need or can do more (e.g., ChEMBL does not seem to have restrictions), change the value. You can also have several instance but again, if you send too many requests at the same, your future requests may be retricted. Currently implemented for REST only
All instances have an attribute called
loggingthat is an instanceof theloggingmodule. It can be used to print information, warning, error messages:self.logging.info("informative message") self.logging.warning("warning message") self.logging.error("error message")
The attribute
debugLevelcan be used to set the behaviour of the logging messages. If the argument verbose is True, the debugLebel is set to INFO. If verbose if False, the debugLevel is set to WARNING. However, you can use thedebugLevelattribute to change it to one of DEBUG, INFO, WARNING, ERROR, CRITICAL. debugLevel=WARNING means that only WARNING, ERROR and CRITICAL messages are shown.- compounds2accession(compounds)[source]¶
For each compound, identifies the target and corresponding UniProt accession number
This is not part of ChEMBL API
# we recommend to use cache if you use this method regularly c = Chembl(cache=True) drugs = c.get_approved_drugs() # to speed up example drugs = drugs[0:20] IDs = [x['molecule_chembl_id] for x in drugs] c.compounds2accession(IDs)
- get_ATC(limit=20, offset=0, filters=None)[source]¶
WHO ATC Classification for drugs
c.get_atc() c[‘atc’]
Note
get_molecule returns ‘molecules’ and likewise all methods return a dictionary whose key is the plural of the method name. This is quite consistent through the API except for that one because it is an acronym
- get_activity(query=None, limit=20, offset=0, filters=None)[source]¶
Activity values recorded in an Assay
- get_approved_drugs(max_phase=4, maxdrugs=1000000)[source]¶
Return all approved drugs
- Parameters
max_phase – 4 by default for approved drugs.
- get_assay(query=None, limit=20, offset=0, filters=None)[source]¶
Assay details as reported in source Document/Dataset
>>> c.get_assay("CHEMBL1217643")
- get_biotherapeutic(limit=20, offset=0, filters=None)[source]¶
Biotherapeutic molecules, which includes HELM notation and sequence data
- get_chembl_id_lookup(query=None, limit=20, offset=0, filters=None)[source]¶
Look up ChEMBL Id entity type
- get_compound_record(query=None, limit=20, offset=0, filters=None)[source]¶
Occurence of a given compound in a spcecific document
- get_compound_structural_alert(query=None, limit=20, offset=0, filters=None)[source]¶
Indicates certain anomaly in compound structure
- get_document(query=None, limit=20, offset=0, filters=None)[source]¶
Document/Dataset from which Assays have been extracted
- get_document_similarity(query=None, limit=20, offset=0, filters=None)[source]¶
Provides documents similar to a given one
- get_document_term(query=None, limit=20, offset=0, filters=None)[source]¶
Provides keywords extracted from a document using the TextRank algorithm
- get_drug(query=None, limit=20, offset=0, filters=None)[source]¶
Approved drugs information, icluding (but not limited to) applicants, patent numbers and research codes
- get_drug_indication(query=None, limit=20, offset=0, filters=None)[source]¶
Joins drugs with diseases providing references to relevant sources
- get_image(query, dimensions=500, format='png', save=True, view=True, engine='indigo')[source]¶
Get the image of a given compound in PNG png format.
- Parameters
- Returns
the path (list of paths) used to save the figure (figures) (different from Chembl API)
>>> from pylab import imread, imshow >>> from bioservices import * >>> s = ChEMBL(verbose=False) >>> res = s.get_image(31863) >>> imshow(imread(res['filenames'][0]))
Todo
ignorecoords option
- get_mechanism(query=None, limit=20, offset=0, filters=None)[source]¶
Mechanism of action information for FDA-approved drugs
- get_metabolism(query=None, limit=20, offset=0, filters=None)[source]¶
Metabolic pathways with references
- get_molecule(query=None, limit=20, offset=0, filters=None)[source]¶
Returns some molecules
- Parameters
limit – number of molecules to retrieve
offset – molecules to ignore before retrieving molecules.
- Returns
a dictionary with keys page_meta and molecules.
There are 1,800,000 molecules (Jan 2019). You can only retrieve 1,000 molecule at most using the limit parameter. With a loop you can retrieve molecules in some range.
c.get_molecule('QFFGVLORLPOAEC-SNVBAGLBSA-N') c.get_molecule("CC(=O)Oc1ccccc1C(=O)O")
- get_molecule_form(query=None, limit=20, offset=0, filters=None)[source]¶
Relationships between molecule parents and salts
>>> s.get_molecule_form("CHEMBL2")['molecule_forms'] [{'is_parent': 'True', 'molecule_chembl_id': 'CHEMBL2', 'parent_chembl_id': 'CHEMBL2'}, {'is_parent': 'False', 'molecule_chembl_id': 'CHEMBL1558', 'parent_chembl_id': 'CHEMBL2'}, {'is_parent': 'False', 'molecule_chembl_id': 'CHEMBL1347191', 'parent_chembl_id': 'CHEMBL2'}]
- get_protein_class(query=None, limit=20, offset=0, filters=None)[source]¶
Protein family classification of TargetComponents
- get_similarity(structure, similarity=80, limit=20, offset=0, filters=None)[source]¶
Molecule similarity search
- Parameters
structure – provide a valid / existing substructure in SMILE format to look for in all molecules:
similarity – must be an integer greater than 70 and less than 100
- Returns
list of molecules corresponding to the search
>>> from bioservices import ChEMBL >>> c = ChEMBL() >>> res = c.get_similarity("CC(=O)Oc1ccccc1C(=O)O", 80) >>> res['molecules']
Here are more examples:
# Similarity (80% cut off) search for against ChEMBL using # aspirin SMILES string c.get_similarity("CC(=O)Oc1ccccc1C(=O)O") # 80 by default # Similarity (80% cut off) search for against ChEMBL using # aspirin CHEMBL_ID c.get_similarity("CHEMBL25") # Similarity (80% cut off) search for against ChEMBL # using aspirin InChI Key c.get_similarity("BSYNRYMUTXBXSQ-UHFFFAOYSA-N")
The ‘Substructure’ and ‘Similarity’ web service resources allow for the chemical content of ChEMBL to be searched. Similar to the other resources, these search based resources except filtering, paging and ordering arguments. These methods accept SMILES, InChI Key and molecule ChEMBL_ID as arguments and in the case of similarity searches an additional identity cut-off is needed. Some example molecule searches are provided in the table below.
Searching with InChI key is only possible for InChI keys found in the ChEMBL database. The system does not try and convert InChI key to a chemical representation.
- get_status()[source]¶
Return version of the DB and number of entries
Returns the number of entries for activities, compound_records, distinct_compounds (molecule), publications (document), targets, etc…
See also
- get_status_resources()[source]¶
Return number of entries for all resources
Note
not in the ChEMBL API.
Changed in version 1.7.3: (removed target_prediction and document_term)
- get_substructure(structure, limit=20, offset=0, filters=None)[source]¶
Molecule substructure search
- Parameters
structure – provide a valid / existing substructure in SMILE format to look for in all molecules:
- Returns
list of molecules corresponding to the search
>>> from bioservices import ChEMBL >>> c = ChEMBL() >>> res = c.get_substructure("CC(=O)Oc1ccccc1C(=O)O")
Other examples:
# Substructure search for against ChEMBL using aspirin # SMILES string c.get_substructure("CC(=O)Oc1ccccc1C(=O)O") # Substructure search for against ChEMBL using aspirin # CHEMBL_ID c.get_substructure("CHEMBL25") # Substructure search for against ChEMBL using aspirin # InChIKey c.get_substructure("BSYNRYMUTXBXSQ-UHFFFAOYSA-N")
The ‘Substructure’ and ‘Similarity’ web service resources allow for the chemical content of ChEMBL to be searched. Similar to the other resources, these search based resources except filtering, paging and ordering arguments. These methods accept SMILES, InChI Key and molecule ChEMBL_ID as arguments and in the case of similarity searches an additional identity cut-off is needed. Some example molecule searches are provided in the table below.
Searching with InChI key is only possible for InChI keys found in the ChEMBL database. The system does not try and convert InChI key to a chemical representation.
- get_target(query=None, limit=20, offset=0, filters=None)[source]¶
Targets (protein and non-protein) defined in Assay
>>> from bioservices import * >>> s = ChEMBL(verbose=False) >>> resjson = s.get_targetd('CHEMBL240')
- get_target_component(query=None, limit=20, offset=0, filters=None)[source]¶
Target sequence information (A Target may have 1 or more sequences)
res = c.get_target_component(1) res['sequence']
- get_target_prediction(query=None, limit=20, offset=0, filters=None)[source]¶
Predictied binding of a molecule to a given biological target
>>> res = c.get_target_prediction(1) >>> res['molecule_chembl_id'] 'CHEMBL2'
- get_target_relation(query=None, limit=20, offset=0, filters=None)[source]¶
Describes relations between targets
>>> c.get_target_relation('CHEMBL261') {'related_target_chembl_id': 'CHEMBL2095180', 'relationship': 'SUBSET OF', 'target_chembl_id': 'CHEMBL261'}
- get_tissue(query=None, limit=20, offset=0, filters=None)[source]¶
Tissue classification
c.get_tissue(filters=[‘pref_name__contains=cervix’])
- order_by(data, name, ascending=True)[source]¶
Ordering data
we use same API as ChEMBL API using the double underscore to indicate a hierarchy in the dictionary. So to access to d[‘a’][‘b’], we use a__b as the input name parameter. We only allows 3 levels e.g., a__b__c
data = c.get_molecules() data1 = c.order_by(data, 'molecule_chembl_id') data2 = c.order_by(data, 'molecule_properties__alogp')
Note
the ChEMBL API allows for ordering but we do not use that API. Instead, we provide this generic function.
8.10. COG¶
Interface to some part of the UniProt web service
What is COG service?
Database of Clusters of Orthologous Genes (COGs)
—From COG web site, Jan 2021
- class COG(verbose=False, cache=False)[source]¶
Interface to the COG service
from bioservices import COG c = COG() cogs = c.get_all_cogs() # This is a pandas dataframe
Constructor
- get_cog_definition_by_name(cog)[source]¶
Get specific COG Definitions by name: Thiamin-binding stress-response protein YqgV, UPF0045 family
- get_cogs(page=1)[source]¶
Get COGs. Unfortunately, the API sends 10 COGS at a tine given a specific page.
The dictionary returned contains the results, count, previous and next page.
- get_cogs_by_id_and_category(cog_id, category)[source]¶
Filter COGs by COG id and Taxonomy Categories: COG0004 and CYANOBACTERIA
8.11. ENA¶
This module provides a class ENA
What is ENA
The European Nucleotide Archive (ENA) provides a comprehensive record of the world’s nucleotide sequencing information, covering raw sequencing data, sequence assembly information and functional annotation.
—From ENA web page Jan 2016
New in version 1.4.4.
- class ENA(verbose=False, cache=False)[source]¶
Interface to ChEMBL
Here is a quick example to retrieve a target given its ChEMBL Id
>>> from bioservices import ENQ >>> s = ENA(verbose=False)
Retrieve read domain metadata in XML format:
print(e.get_data('ERA000092', 'xml'))
Retrieve assemble and annotated sequences in fasta format:
print(e.get_data('A00145', 'fasta'))
The range parameter can be used in combination to retrieve a subsequence from sequence entry A00145 from bases 3 to 63 using
e.get_data('A00145', 'fasta', fasta_range=[3,63])
Retrieve assembled and annotated subsequences in HTML format (same as above but in HTML page).
e.view_data(‘A00145’)
Retrieve expanded CON records:
To retrieve expanded CON records use the expanded=true parameter. For example, the expanded CON entry AL513382 in flat file format can be i obtained as follows:
e.get_data('AL513382', frmt='text', expanded=True)
Expanded CON records are different from CON records in two ways. Firstly, the expanded CON records contain the full sequence in addition to the contig assembly instructions. Secondly, if a CON record contains only source or gap features the expanded CON records will also display all features from the segment records.
Retrieve assembled and annotated sequence header in flat file format
To retrieve assembled and annotated sequence header in flat file format please use the header=true parameter, e.g.:
e.get_data(‘BN000065’, ‘text’, header=True)
Retrieve assembled and annotated sequence records using sequence versions:
e.get_data('AM407889.1', 'fasta') e.get_data('AM407889.2', 'fasta')
Constructor
- Parameters
verbose – set to False to prevent informative messages
- get_data(identifier, frmt, fasta_range=None, expanded=None, header=None, download=None)[source]¶
- :param frmtxml, text, fasta, fastq, html, embl but does depend on the
entry
Example:
get_data(“/AL513382”, “embl”)
ENA API changed in 2020 but we tried to keep the same services in this method.
- url = 'http://www.ebi.ac.uk/ena/browser/api'¶
8.12. EUtils¶
Interface to the EUtils web Service.
What is EUtils ?
The Entrez Programming Utilities (E-utilities) are a set of eight server-side programs that provide a stable interface into the Entrez query and database system at the National Center for Biotechnology Information (NCBI). The E-utilities use a fixed URL syntax that translates a standard set of input parameters into the values necessary for various NCBI software components to search for and retrieve the requested data. The E-utilities are therefore the structured interface to the Entrez system, which currently includes 38 databases covering a variety of biomedical data, including nucleotide and protein sequences, gene records, three-dimensional molecular structures, and the biomedical literature.
—from http://www.ncbi.nlm.nih.gov/books/NBK25497/, March 2013
- class EUtils(verbose=False, email='unknown', cache=False, xmlparser='EUtilsParser')[source]¶
Interface to NCBI Entrez Utilities service
Note
Technical note: the WSDL interface was dropped in july 2015 so we now use the REST service.
Warning
Read the guidelines before sending requests. No more than 3 requests per seconds otherwise your IP may be banned. You should provide your email by filling the
emailso that before being banned, you may be contacted.There are a few methods such as
ELink(),EFetch(). Here is an example on how to useEFetch()method to retrieve the FASTA sequence of a given identifier (34577063):>>> from bioservices import EUtils >>> s = EUtils() >>> print(s.EFetch("protein", "34577063", rettype="fasta")) >gi|34577063|ref|NP_001117.2| adenylosuccinate synthetase isozyme 2 [Homo sapiens] MAFAETYPAASSLPNGDCGRPRARPGGNRVTVVLGAQWGDEGKGKVVDLLAQDADIVCRCQGGNNAGHTV VVDSVEYDFHLLPSGIINPNVTAFIGNGVVIHLPGLFEEAEKNVQKGKGLEGWEKRLIISDRAHIVFDFH QAADGIQEQQRQEQAGKNLGTTKKGIGPVYSSKAARSGLRMCDLVSDFDGFSERFKVLANQYKSIYPTLE IDIEGELQKLKGYMEKIKPMVRDGVYFLYEALHGPPKKILVEGANAALLDIDFGTYPFVTSSNCTVGGVC TGLGMPPQNVGEVYGVVKAYTTRVGIGAFPTEQDNEIGELLQTRGREFGVTTGRKRRCGWLDLVLLKYAH MINGFTALALTKLDILDMFTEIKVGVAYKLDGEIIPHIPANQEVLNKVEVQYKTLPGWNTDISNARAFKE LPVNAQNYVRFIEDELQIPVKWIGVGKSRESMIQLF
Most of the methods take a database name as input. You can obtain the valid list by checking the
databasesattribute.A few functions takes Identifier(s) as input. It could be a list of strings, list of numbers, or a string where identifiers are separated either by comma or spaces.
A few functions take an argument called term. You can use the AND keyword with spaces or + signs as separators:
Correct: term=biomol mrna[properties] AND mouse[organism] Correct: term=biomol+mrna[properties]+AND+mouse[organism]
Other special characters, such as quotation marks (”) or the # symbol used in referring to a query key on the History server, could be represented by their URL encodings (%22 for “; %23 for #) or verbatim .:
Correct: term=#2+AND+"gene in genomic"[properties] Correct: term=%232+AND+%22gene+in+genomic%22[properties]
For information about retmode and retype, please see:
- ECitMatch(bdata, **kargs)[source]¶
- Parameters
bdata –
Citation strings. Each input citation must be represented by a citation string in the following format:
journal_title|year|volume|first_page|author_name|your_key|
Multiple citation strings may be provided by separating the strings with a carriage return character (%0D) or simply \r or \n.
The your_key value is an arbitrary label provided by the user that may serve as a local identifier for the citation, and it will be included in the output.
all spaces must be replaced by + symbols and that citation strings should end with a final vertical bar |.
Only xml supported at the time of this implementation.
from bioservices import EUtils s = EUtils() print(s.ECitMatch("proc+natl+acad+sci+u+s+a|1991|88|3248|mann+bj|Art1|%0Dscience|1987|235|182|palmenberg+ac|Art2|"))
- EFetch(db, id, retmode='text', **kargs)[source]¶
Access to the EFetch E-Utilities
- Parameters
- Returns
depends on retmode parameter.
Note
addition to NCBI: settings rettype to “dict” returns a dictionary
>>> ret = s.EFetch("omim", "269840") --> ZAP70 >>> ret = s.EFetch("taxonomy", "9606", retmode="xml") >>> [x.text for x in ret.getchildren()[0].getchildren() if x.tag=="ScientificName"] ['Homo sapiens'] >>> s = eutils.EUtils() >>> s.EFetch("protein", "34577063", retmode="text", rettype="fasta") >gi|34577063|ref|NP_001117.2| adenylosuccinate synthetase isozyme 2 [Homo sapiens] MAFAETYPAASSLPNGDCGRPRARPGGNRVTVVLGAQWGDEGKGKVVDLLAQDADIVCRCQGGNNAGHTV VVDSVEYDFHLLPSGIINPNVTAFIGNGVVIHLPGLFEEAEKNVQKGKGLEGWEKRLIISDRAHIVFDFH QAADGIQEQQRQEQAGKNLGTTKKGIGPVYSSKAARSGLRMCDLVSDFDGFSERFKVLANQYKSIYPTLE IDIEGELQKLKGYMEKIKPMVRDGVYFLYEALHGPPKKILVEGANAALLDIDFGTYPFVTSSNCTVGGVC TGLGMPPQNVGEVYGVVKAYTTRVGIGAFPTEQDNEIGELLQTRGREFGVTTGRKRRCGWLDLVLLKYAH MINGFTALALTKLDILDMFTEIKVGVAYKLDGEIIPHIPANQEVLNKVEVQYKTLPGWNTDISNARAFKE LPVNAQNYVRFIEDELQIPVKWIGVGKSRESMIQLF
Identifiers could be provided as a single string with comma-separated values, or a list of strings, a list of integers, or just one string or one integer but no mixing of types in the list:
>>> e.EFetch("protein", "352, 234", retmode="text", rettype="fasta") >>> e.EFetch("protein", 352, retmode="text", rettype="fasta") >>> e.EFetch("protein", [352], retmode="text", rettype="fasta") >>> e.EFetch("protein", [352, 234], retmode="text", rettype="fasta")
retmode should be xml or text depending on the database. For instance, xml for pubmed:
>>> e.EFetch("pubmed", "20210808", retmode="xml") >>> e.EFetch('nucleotide', id=15, retmode='xml') >>> e.EFetch('nucleotide', id=15, retmode='text', rettype='fasta') >>> e.EFetch('nucleotide', 'NT_019265', rettype='gb')
Other special characters, such as quotation marks (”) or the # symbol used in referring to a query key on the History server, should be represented by their URL encodings (%22 for “; %23 for #).
A useful command is the following one that allows to get back a GI identifier from its accession, which is common to NCBI/EMBL:
e.EFetch(db="nuccore",id="AP013055", rettype="seqid", retmode="text")
Changed in version 1.5.0: instead of “xml”, retmode can now be set to dict, in which case an XML is retrieved and converted to a dictionary if possible.
- EGQuery(term, **kargs)[source]¶
Provides the number of records retrieved in all Entrez databases by a text query.
- Parameters
term (str) – Entrez text query. Spaces may be replaced by ‘+’ signs. For very long queries (more than several hundred characters long), consider using an HTTP POST call. See the PubMed or Entrez help for information about search field descriptions and tags. Search fields and tags are database specific.
- Returns
returns a json data structure
>>> ret = s.EGQuery("asthma") >>> [(x.DbName, x.Count) for x in ret.eGQueryResult.ResultItem if x.Count!='0'] >>> ret = s.EGQuery("asthma") >>> ret.eGQueryResult.ResultItem[0] {'Count': '115241', 'DbName': 'pmc', 'MenuName': 'PubMed Central', 'Status': 'Ok'}
- EInfo(db=None, **kargs)[source]¶
Provides information about a database (e.g., number of records)
- Parameters
db (str) – target database about which to gather statistics. Value must be a valid Entrez database name. See
databasesor don’t provide any value to obtain the entire list- Returns
a json data structure that depends on the value of
databases(default to json)
>>> all_database_names = s.EInfo() >>> # specific info about one database: >>> ret = s.EInfo("taxonomy") >>> ret[0]['count'] u'1445358' >>> ret = s.EInfo('pubmed') >>> ret[0]['fieldlist'][2]['fullname'] 'Filter'
You can use the retmode parameter to ‘xml’ as well. In that case, you will need a XML parser.
>>> ret = s.EInfo("taxonomy")
Note
Note that the name in the XML or json outputs differ (some have lower cases, some have upper cases). This is inherent to the output of EUtils.
- ELink(db=None, dbfrom=None, id=None, **kargs)[source]¶
The Entrez links utility
Responds to a list of UIDs in a given database with either a list of related UIDs (and relevancy scores) in the same database or a list of linked UIDs in another Entrez database;
- Parameters
db (str) – valid database from which to retrieve UIDs.
dbfrom (str) – Database containing the input UIDs. The value must be a valid database name (default = pubmed). This is the origin database of the link operation. If db and dbfrom are set to the same database value, then ELink will return computational neighbors within that database. Computational neighbors have linknames that begin with dbname_dbname (examples: protein_protein, pcassay_pcassay_activityneighbor).
id (str) – UID list. Either a single UID or a comma-delimited list Limited to 200 Ids
cmd (str) – ELink command mode. The command mode specified which function ELink will perform. Some optional parameters only function for certain values of cmd (see http://www.ncbi.nlm.nih.gov/books/NBK25499/#chapter4.ELink). Examples are neighbor, prlinks.
>>> # Example: Find related articles to PMID 20210808 >>> ret = s.ELink("pubmed", id="20210808", cmd="neighbor_score") >>> ret = s.parse_xml(ret, 'EUtilsParser') >>> ret.eLinkResult.LinkSet.LinkSetDb[0].Link[1] {'Id': '16539535'} >>> s.ELink(dbfrom="nucleotide", db="protein", id="48819,7140345") >>> s.ELink(dbfrom="nucleotide", db="protein", id="48819,7140345") >>> s.ELink(dbfrom='nuccore', id='21614549,219152114', cmd='ncheck')
Convert GI number to Taxon identifiers:
>>> s.ELink(dbfrom='nuccore', db="taxonomy", id='21614549,219152114')
- EPost(db, id, **kargs)[source]¶
Accepts a list of UIDs from a given database,
stores the set on the History Server, and responds with a query key and web environment for the uploaded dataset.
- Parameters
db (str) – a valid database
id – list of strings of strings
- Returns
a dictionary with a Web Environment string and a QueryKey to be re-used in another EUtils.
- ESearch(db, term, **kargs)[source]¶
Responds to a query in a given database
The response can be used later in ESummary, EFetch or ELink, along with the term translations of the query.
- Parameters
db – a valid database
term – an Entrez text query
Note
see
_get_esearch_params()for the list of valid parameters.>>> ret = e.ESearch('protein', 'human', RetMax=5) >>> ret = e.ESearch('taxonomy', 'Staphylococcus aureus[all names]') >>> ret = e.ESearch('pubmed', "cokelaer AND BioServices") >>> ret = e.ESearch('protein', '15718680') >>> # Let us show the first pubmed identifier in a browser >>> identifiers = e.pubmed(ret['idlist'][0])
More complex requests can be used. We will not cover all the possiblities (see the NCBI website). Here is an example to tune the search term to look into PubMed for the journal PNAS Volume 16, and retrieve.:
>>> e.ESearch("pubmed", "PNAS[ta] AND 16[vi]")
You can then look more closely at a specific identifier using EFetch:
>>> e = EFetch("pubmed") >>> e.Efetch(identifiers)
Note
valid parameters can be found by calling
_get_esearch_params()
- ESpell(db, term, **kargs)[source]¶
Retrieve spelling suggestions for a text query in a given database.
- Parameters
>>> ret = e.ESpell(db="pubmed", term="aasthma+OR+alergy") >>> ret = ret['eSpellResult'] >>> ret['Query'] 'asthmaa OR alergies' >>> ret['CorrectedQuery'] 'asthma or allergy' >>> ret = e.ESpell(db="pubmed", term="biosservices") >>> ret = ret['eSpellResult'] >>> ret['CorrectedQuery'] bioservices
- ESummary(db, id=None, **kargs)[source]¶
Returns document summaries for a list of input UIDs
- Parameters
db – a valid database
id (str) – list of identifiers (or string comma separated). all of the UIDs must be from the database specified by db. Limited to 200 identifiers
>>> from bioservices import * >>> s = EUtils() >>> ret = s.ESummary("snp","7535") >>> ret = s.ESummary("snp","7535,7530") >>> ret = s.ESummary("taxonomy", "9606,9913")
>>> proteins = e.ESearch("protein", "bacteriorhodopsin", retmax=20) >>> ret = e.ESummary("protein", 449301857) >>> ret['result']['449301857']['extra'] 'gi|449301857|gb|EMC97866.1||gnl|WGS:AEIF|BAUCODRAFT_31870'
- property databases¶
Returns list of valid databases
- email¶
fill this with your email address
- class EUtilsParser(xml)[source]¶
Convert xml returned by EUtils into a structure easier to manipulate
Used by
EUtils.EGQuery(),EUtils.ELink().
8.13. GeneProf¶
Currently removed from the main API from version 1.6.0 onwards. You can still get the code in earlier version or in the github repository in the attic/ directory
8.14. QuickGO¶
Interface to the quickGO interface
What is quickGO
“QuickGO is a fast web-based browser for Gene Ontology terms and annotations, which is provided by the UniProt-GOA project at the EBI. “
—from QuickGO home page, Dec 2012
- class QuickGO(verbose=False, cache=False)[source]¶
Interface to the QuickGO service
Retrieve information given a GO identifier:
>>> from bioservices import QuickGO >>> go = QuickGO() >>> res = go.get_go_terms("GO:0003824")
Changed in version we: use the new QuickGO API since version 1.5.0 To use the old API, please use version of bioservices below 1.5
Constructor
- Parameters
verbose (bool) – print informative messages.
- Annotation(assignedBy=None, includeFields=None, limit=100, page=1, aspect=None, reference=None, geneProductId=None, evidenceCode=None, goId=None, qualifier=None, withFrom=None, taxonId=None, taxonUsage=None, goUsage=None, goUsageRelationships=None, evidenceCodeUsage=None, evidenceCodeUsageRelationships=None, geneProductType=None, targetSet=None, geneProductSubset=None, extension=None)[source]¶
Calling the Annotation service
Changed in version 1.4.18: due to service API changes, we refactored this method completely
- Parameters
assignedBy (str) – The database from which this annotation originates. Accepts comma separated values.E.g., BHF-UCL,Ensembl.
includeFields (str) – Optional fields retrieved from external services. Accepts comma separated values. accepted values: goName, taxonName, name, synonyms.
limit (int) – download limit (number of lines) (default 10,000 rows, which may not be sufficient for the data set that you are downloading. To bypass this default, and return the entire data set, specify a limit of -1).
page (int) – results may be stored on several pages. You must provide this number. There is no way to retrieve more than 100 results without calling this function several times chanding this parameter (default to 1).
aspect (char) – use this to limit the annotations returned to a specific ontology or ontologies (Molecular Function, Biological Process or Cellular Component). The valid character can be F,P,C.
reference (str) – PubMed or GO reference supporting annotation. Can refer to a specific reference identifier or category (for category level, use * after ref type). Can be ‘PUBMED:*’, ‘GO_REF:0000002’.
geneProductId (str) – The id of the gene product annotated with the GO term. Accepts comma separated values.E.g., URS00000064B1_559292.
evidenceCode (str) – Evidence code indicating how the annotation is supported. Accepts comma separated values. E.g., ECO:0000255.
goId (str) – The GO id of an annotation. Accepts comma separated values. E.g., GO:0070125.
qualifier (str) – Aids the interpretation of an annotation. Accepts comma separated values. E.g., enables,involved_in.
withFrom (str) – Additional ids for an annotation. Accepts comma separated values. E.g., P63328.
taxonId (str) – The taxonomic id of the species encoding the gene product associated to an annotation. Accepts comma separated values. E.g., 1310605.
taxonUsage (str) – Indicates how the taxonomic ids within the annotations should be used. E.g., exact.
goUsage (str) – Indicates how the GO terms within the annotations should be used. Used in conjunction with ‘goUsageRelationships’ filter. E.g., descendants.
goUsageRelationships (str) – The relationship between the ‘goId’ values found within the annotations. Allows comma separated values. E.g., is_a,part_of.
evidenceCodeUsage (str) – Indicates how the evidence code terms within the annotations should be used. Is used in conjunction with ‘evidenceCodeUsageRelationships’ filter. E.g., descendants, exact<F12>
evidenceCodeUsageRelationships (str) – The relationship between the provided ‘evidenceCode’ identifiers. Allows comma separated values. E.g., is_a,part_of.
geneProductType (str) – The type of gene product. Accepts comma separated values. E.g., protein,RNA. can be protein, RNA and/or complex
targetSet (str) – Gene product set. Accepts comma separated values. E.g., KRUK,BHF-UCL,Exosome.
geneProductSubset (str) – A database that provides a set of gene products. Accepts comma separated values. E.g., TrEMBL.
extension (str) – Extensions to annotations, where each extension can be: EXTENSION(DB:ID) / EXTENSION(DB) / EXTENSION.
- Returns
a dictionary
>>> print(s.Annotation(protein='P12345', frmt='tsv', col="ref,evidence", ... reference='PMID:*')) >>> print(s.Annotation(protein='P12345,Q4VCS5', frmt='tsv', ... col="ref,evidence",reference='PMID:,Reactome:'))
- Annotation_from_goid(goId, max_number_of_pages=25, **kargs)[source]¶
Returns a DataFrame containing annotation on a given GO identifier
- Parameters
protein (str) – a GO identifier
- Returns
all outputs are stored into a Pandas.DataFrame data structure.
All parameters from
 are also valid except format that
is set to tsv and cols that is made of all possible column names.
are also valid except format that
is set to tsv and cols that is made of all possible column names.
- gene_product_search(query, taxonID=None, page=1, limit=100, type=None, dbSubSet=None, proteome=None)[source]¶
- get_go_chart(query)[source]¶
res = go.get_chart("GO:0022804") with open("temp.png", "wb") as fout: fout.write(res)
- get_go_paths(_from, _to, relations='is_a,part_of,occurs_in,regulates')[source]¶
Retrieves the paths between two specified sets of ontology terms. Each path is formed from a list of (term, relationship, term) triples.
paths = go.go_terms_path(”GO:0005215”, “GO:0003674”) # First path is found as the first item in the “results” paths[“results”][0]
8.15. Kegg¶
This module provides a class KEGG to access to the
REST KEGG interface. There are additional methods and functionalities added by
BioServices.
Note
a previous imterface to the KEGG WSDL service was designed but the WSDL closed in Dec 2012.
What is KEGG ?
- URL
- REST
- weblink
- dbentries
“KEGG is a database resource for understanding high-level functions and utilities of the biological system, such as the cell, the organism and the ecosystem, from molecular-level information, especially large-scale molecular datasets generated by genome sequencing and other high-throughput experimental technologies (See Release notes for new and updated features). “
—KEGG home page, Jan 2013
8.15.1. Some terminology¶
The following list is a simplified list of terminology taken from KEGG API pages.
organisms (org) are made of a three-letter (or four-letter) code (e.g., hsa stands for Human Sapiens) used in KEGG (see
organismIds).db is a database name. See
databasesattribute and KEGG Databases Names and Abbreviations section.entry_id is a unique identifier. It is a combination of the database name and the identifier of an entry joined by a colon sign (e.g. ‘embl:J00231’).
Here are some examples of entry Ids:
genes_id: A KEGG organism and a gene name (e.g. ‘eco:b0001’).
enzyme_id: ‘ec’ and an enzyme code. (e.g. ‘ec:1.1.1.1’). See
enzymeIds.compound_id: ‘cpd’ and a compound number (e.g. ‘cpd:C00158’). Some compounds also have ‘glycan_id’ and both IDs are accepted and converted internally. See
compoundIds.drug_id: ‘dr’ and a drug number (e.g. ‘dr:D00201’). See
drugIds.glycan_id: ‘gl’ and a glycan number (e.g.
‘gl:G00050’). Some glycans also have ‘compound_id’ and both IDs are accepted and converted internally. see
glycanIdsattribute.reaction_id: ‘rn’ and a reaction number (e.g.
‘rn:R00959’ is a reaction which catalyze cpd:C00103 into cpd:C00668). See
reactionIdsattribute.pathway_id: ‘path’ and a pathway number. Pathway numbers prefixed by ‘map’ specify the reference pathway and pathways prefixed by a KEGG organism specify pathways specific to the organism (e.g. ‘path:map00020’, ‘path:eco00020’). See
pathwayIdsattribute.motif_id: a motif database names (‘ps’ for prosite, ‘bl’ for blocks, ‘pr’ for prints, ‘pd’ for prodom, and ‘pf’ for pfam) and a motif entry name. (e.g. ‘pf:DnaJ’ means a Pfam database entry ‘DnaJ’).
ko_id: identifier made of ‘ko’ and a ko number (e.g. ‘ko:K02598’). See
koIdsattribute.
8.15.2. KEGG Databases Names and Abbreviations¶
Here is a list of databases used in KEGG API with their name and abbreviation:
Database Name |
Abbrev |
kid |
|---|---|---|
pathway |
path |
map number |
brite |
br |
br number |
module |
md |
M number |
disease |
ds |
H number |
drug |
dr |
D number |
environ |
ev |
E number |
orthology |
ko |
K number |
genome |
genome |
T number |
genomes |
gn |
T number |
genes |
||
ligand |
ligand |
|
compound |
cpd |
C number |
glycan |
gl |
G number |
reaction |
rn |
R number |
rpair |
rp |
RP number |
rclass |
rc |
RC number |
enzyme |
ec |
8.15.3. Database Entries¶
Database entries can be written in on of the following ways:
<dbentries> = <dbentry>1[+<dbentry>2...]
<dbentry> = <db:entry> | <kid> | <org:gene>
Each database entry is identified by:
db:entry
where “db” is the database name or its abbreviation shown above and “entry” is the entry name or the accession number that is uniquely assigned within the database. In reality “db” may be omitted, for the entry name called the KEGG object identifier (kid) is unique across KEGG.:
kid = database-dependent prefix + five-digit number
In the KEGG GENES database the db:entry combination must be specified. This is more specifically written as:
org:gene
where “org” is the three- or four-letter KEGG organism code or the T number genome identifier and “gene” is the gene identifier, usually locus_tag or ncbi GeneID, or the primary gene name.
- class KEGG(verbose=False, cache=False)[source]¶
Interface to the KEGG service
This class provides an interface to the KEGG REST API. The weblink tools are partially accesible. All dbentries can be parsed into dictionaries using the
KEGGParserHere are some examples. In order to retrieve the entry of the gene identifier 7535 of the hsa organism, type:
from bioservices import KEGG s = KEGG() print(s.get("hsa:7535"))
The output is the raw ouput sent by KEGG API. See
KEGGParserto parse this output.See also
The Database Entries to know more about the db entries format.
Another example here below shows how to print the list of pathways of the human organism:
print(s.list("pathway", organism="hsa"))
Further post processing would allow you to retrieve the pathway Ids. However, we provide additional functions to the KEGG API so the previous code and post processing to extract the pathway Ids can be written as:
s.organism = "hsa" s.pathwayIds
and similarly you can get all
databases()output and database Ids easily. For example, for the reaction database:s.reaction # equivalent to s.list("reaction") s.reactionIds
Other methods of interest are
conv(),find(),get().Constructor
- Parameters
verbose (bool) – prints informative messages
- Tnumber2code(Tnumber)[source]¶
Converts organism T number to its code
>>> from bioservices import KEGG >>> s = KEGG() >>> s.Tnumber2code("T01001") 'hsa'
- code2Tnumber(code)[source]¶
Converts organism code to its T number
>>> from bioservices import KEGG >>> s = KEGG() >>> s.code2Tnumber("hsa") 'T01001'
- conv(target, source)[source]¶
convert KEGG identifiers to/from outside identifiers
- Parameters
- Returns
a dictionary with keys being the source and values being the target.
Here are the rules to set the target and source parameters.
If the second argument is not a dbentries, source and target parameters can be of two types:
gene identifiers. If the target is a KEGG Id, then the source must be one of ncbi-gi, ncbi-geneid or uniprot.
Note
source and target can be swapped.
chemical substance identifiers. If the target is one of the following kegg database: drug, compound, glycan then the source must be one of pubchem or chebi.
Note
again, source and target can be swapped
If the second argument is a dbentries, it can be again of two types:
gene identifiers. The database used can be one ncbi-gi, ncbi-geneid, uniprot or any KEGG organism
chemical substance identifiers. The database used can be one of drug, compound, glycan, pubchem or chebi only.
Note
if the second argument is a dbentries, target and dbentries cannot be swapped.
# conversion from NCBI GeneID to KEGG ID for E. coli genes conv("eco","ncbi-geneid") # inverse of the above example conv("eco","ncbi-geneid") #conversion from KEGG ID to NCBI GI conv("ncbi-gi","hsa:10458+ece:Z5100")
To make it clear by taking another example, you can either convert an entire database to another (e.g., from uniprot to KEGG Id all human gene IDs):
uniprot_ids, kegg_ids = s.conv("hsa", "uniprot")
or a subset by providing a valid dbentries:
s.conv("hsa","up:Q9BV86+")
Warning
dbentries are not check and are supposed to be correct. See
check_idbentries()to help you checking a dbentries.Warning
call to this function may be long. conv(“hsa”, “uniprot”) takes a minute suprinsingly, conv(“uniprot”, “hsa”) takes just a few seconds.
Changed in version 1.1: the output is now a dictionary, not a list of tuples
- property databases¶
Returns list of valid KEGG databases.
- dbinfo(database='kegg')[source]¶
Displays the current statistics of a given database
- Parameters
database (str) – can be one of: kegg (default), brite, module, disease, drug, environ, ko, genome, compound, glycan, reaction, rpair, rclass, enzyme, genomes, genes, ligand or any
organismIds.
from bioservices import KEGG s = KEGG() s.dbinfo("hsa") # human organism s.dbinfo("T01001") # same as above s.dbinfo("pathway")
Changed in version 1.4.1: renamed info method into
dbinfo(), which clashes with Logging framework info() method.
- entry(dbentries)[source]¶
Retrieve entry
There is a weblink service (see http://www.genome.jp/kegg/rest/weblink.html) Since it is equivalent to
get(), we do not implement it for now
- find(database, query, option=None)[source]¶
finds entries with matching query keywords or other query data in a given database
- Parameters
database (str) – can be one of pathway, module, disease, drug, environ, ko, genome, compound, glycan, reaction, rpair, rclass, enzyme, genes, ligand or an organism code (see
organismIdsattributes) or T number (seeorganismTnumbersattribute).query (str) – See examples
option (str) – If option provided, database can be only ‘compound’ or ‘drug’. Option can be ‘formula’, ‘exact_mass’ or ‘mol_weight’
Note
Keyword search against brite is not supported. Use /list/brite to retrieve a short list.
# search for pathways that contain Viral in the definition s.find("pathway", "Viral") # for keywords "shiga" and "toxin" s.find("genes", "shiga+toxin") # for keywords "shiga toxin" s.find("genes", ""shiga toxin") # for chemical formula "C7H10O5" s.find("compound", "C7H10O5", "formula") # for chemical formula containing "O5" and "C7" s.find("compound", "O5C7","formula") # for 174.045 =< exact mass < 174.055 s.find("compound", "174.05","exact_mass") # for 300 =< molecular weight =< 310 s.find("compound", "300-310","mol_weight")
- get(dbentries, option=None, parse=False)[source]¶
Retrieves given database entries
- param str dbentries
KEGG database entries involving the following database: pathway, brite, module, disease, drug, environ, ko, genome compound, glycan, reaction, rpair, rclass, enzyme or any organism using the KEGG organism code (see
organismIdsattributes) or T number (seeorganismTnumbersattribute).- param str option
one of: aaseq, ntseq, mol, kcf, image, kgml
Note
- you can add the option at the end of dbentries in which case
the parameter option must not be used (see example)
from bioservices import KEGG s = KEGG() # retrieves a compound entry and a glycan entry s.get("cpd:C01290+gl:G00092") # same as above s.get("C01290+G00092") # retrieves a human gene entry and an E.coli O157 gene entry s.get("hsa:10458+ece:Z5100") # retrieves amino acid sequences of a human gene and an E.coli O157 gene s.get("hsa:10458+ece:Z5100/aaseq") # retrieves the image file of a pathway map s.get("hsa05130/image") # same as above s.get("hsa05130", "image")
Another example here below shows how to save the image of a given pathway:
res = s.get("hsa05130/image") # same as : res = s.get("hsa05130","image") f = open("test.png", "w") f.write(res) f.close()
Note
The input is limited up to 10 entries (KEGG restriction).
- get_pathway_by_gene(gene, organism)[source]¶
Search for pathways that contain a specific gene
- Parameters
- Returns
list of pathway Ids that contain the gene
>>> s.get_pathway_by_gene("7535", "hsa") ['path:hsa04064', 'path:hsa04650', 'path:hsa04660', 'path:hsa05340']
- isOrganism(org)[source]¶
Checks if org is a KEGG organism
- Parameters
org (str) –
- Returns
True if org is in the KEGG organism list (code or Tnumber)
>>> from bioservices import KEGG >>> s = KEGG() >>> s.isOrganism("hsa") True
- link(target, source)[source]¶
Find related entries by using database cross-references
- Parameters
The valid list of databases is pathway, brite, module, disease, drug, environ, ko, genome, compound, glycan, reaction, rpair, rclass, enzyme
# KEGG pathways linked from each of the human genes s.link("pathway", "hsa") # human genes linked from each of the KEGG pathways s.link("hsa", "pathway") # KEGG pathways linked from a human gene and an E. coli O157 gene. s.link("pathway", "hsa:10458+ece:Z5100")
- list(query, organism=None)[source]¶
Returns a list of entry identifiers and associated definition for a given database or a given set of database entries
- Parameters
query (str) – can be one of pathway, brite, module, disease, drug, environ, ko, genome, compound, glycan, reaction, rpair, rclass, enzyme, organism or an organism from the
organismIdsattribute or a valid dbentry (see below). If a dbentry query is provided, organism should not be used!organism (str) – a valid organism identifier that can be provided. If so, database can be only “pathway” or “module”. If not provided, the default value is chosen (
organism)
- Returns
A string with a structure that depends on the query
Here is an example that shows how to extract the pathways IDs related to the hsa organism:
>>> s = KEGG() >>> res = s.list("pathway", organism="hsa") >>> pathways = [x.split()[0] for x in res.strip().split("\n")] >>> len(pathways) # as of Dec 2012 261
Note, however, that there are convenient aliases to some of the databases. For instance, the pathway Ids can also be retrieved as a list from the
pathwayIdsattribute (after defining theorganismattribute).Note
If you set the query to a valid organism, then the second argument rganism is irrelevant and ignored.
Note
If the query is not a database or an organism, it is supposed to be a valid dbentries string and the maximum number of entries is 100.
Other examples:
s.list("pathway") # returns the list of reference pathways s.list("pathway", "hsa") # returns the list of human pathways s.list("organism") # returns the list of KEGG organisms with taxonomic classification s.list("hsa") # returns the entire list of human genes s.list("T01001") # same as above s.list("hsa:10458+ece:Z5100") # returns the list of a human gene and an E.coli O157 gene s.list("cpd:C01290+gl:G00092")# returns the list of a compound entry and a glycan entry s.list("C01290+G00092") # same as above
- lookfor_organism(query)[source]¶
Look for a specific organism
- Parameters
query (str) – your search term. upper and lower cases are ignored
- Returns
a list of definition that matches the query
- lookfor_pathway(query)[source]¶
Look for a specific pathway
- Parameters
query (str) – your search term. upper and lower cases are ignored
- Returns
a list of definition that matches the query
- property moduleIds¶
returns list of module Ids for the default organism.
organismmust be set.s = KEGG() s.organism = "hsa" s.moduleIds
- property organism¶
returns the current default organism
- property organismIds¶
Returns list of organism Ids
- parse(entry)[source]¶
See
KEGGParserfor detailsParse entry returned by
get()k = KEGG() res = k.get("hsa04150") d = k.parse(res)
- parse_kgml_pathway(pathwayId, res=None)[source]¶
Parse the pathway in KGML format and returns a dictionary (relations and entries)
- Parameters
- Returns
a dictionary with relations and entries as keys. Values of relations is a list of relations, each relation being dictionary with entry1, entry2, link, value, name. The list os entries is a list of dictionary as well. Entry contains contains more details about the entry found in the relation. See example below for details.
>>> res = s.parse_kgml_pathway("hsa04660") >>> set([x['name'] for x in res['relations']]) >>> res['relations'][-1] {'entry1': u'15', 'entry2': u'13', 'link': u'PPrel', 'name': u'phosphorylation', 'value': u'+p'} >>> set([x['link'] for x in res['relations']]) set([u'PPrel', u'PCrel']) >>> # get information about an entry : >>> res['entries'][4]
See also
- pathway2sif(pathwayId, uniprot=True)[source]¶
Extract protein-protein interaction from KEGG pathway to a SIF format
Warning
experimental Not tested on all pathway. should be move to another package such as cellnopt
- Parameters
- Returns
a list of relations (A 1 B) for activation and (A -1 B) for inhibitions
This is longish due to the conversion from KEGGIds to UniProt.
This method can be useful to provide prior knowledge network to software such as CellNOpt (see http://www.cellnopt.org)
- property pathwayIds¶
returns list of pathway Ids for the default organism.
organismmust be set.s = KEGG() s.organism = "hsa" s.pathwayIds
- property reactionIds¶
returns list of reaction Ids
- save_pathway(pathId, filename, scale=None, keggid={}, params={})[source]¶
Save KEGG pathway in PNG format
- Parameters
pathId – a valid pathway identifier
filename (str) – output PNG file
params – valid kegg params expected
- show_module(modId)[source]¶
Show a given module inside a web browser
- Parameters
modId (str) – a valid module Id. See
moduleIds()
Validity of modId is not checked but if wrong the URL will not open a proper web page.
- show_pathway(pathId, scale=None, dcolor='pink', keggid={}, show=True)[source]¶
Show a given pathway inside a web browser
- Parameters
pathId (str) – a valid pathway Id. See
pathwayIds()scale (int) – you can scale the image with a value between 0 and 100
dcolor (str) – set the default background color of nodes
keggid (dict) – set color of entries contained in the pathway as key/value pairs; can also be a list, in which case all nodes have the same default color (red)
Note
if scale is provided, dcolor and keggid are ignored.
# show a pathway in the browser s.show_pathway("path:hsa05416", scale=50) # Same as above but also highlights some KEGG Ids (red for all) s.show_pathway("path:hsa05416", dcolor="white", keggid=['1525', '1604', '2534']) # You can refine the colors using a dictionary: s.show_pathway("path:hsa05416", dcolor="white", keggid={'1525':'yellow,red', '1604':'blue,green', '2534':"blue"})
- class KEGGParser(verbose=False)[source]¶
This is an extension of the
KEGGclass to ease parsing of dbentriesThis class provides a generic method
parse()that will read the output of a dbentry returned byKEGG.get()and converts it into a dictionary ready to use.The
parse()method parses any entry. It can be a pathway, a gene, a compound…from bioservices import * s = KEGG() # Retrieve a KEGG entry res = s.get("hsa04150") # parse it d = s.parse(res)
As a pedagogical example, you can then further process this dictionary. Here below, we convert the gene Ids found in the pathway into UniProt Ids:
# Get the KEGG Ids in the pathway kegg_geneIds = [x for x in d['GENE']] # Convert them db_up, db_kegg = s.conv("hsa", "uniprot") # Get the corresponding uniprot Ids indices = [db_kegg.index("hsa:%s" % x ) for x in kegg_geneIds] uniprot_geneIds = [db_up[x] for x in indices]
However, you could also have done it simply as follows:
kegg_geneIds = [x for x in d['gene']] uprot_geneIds = [s.parse(s.get("hsa:"+str(e)))['DBLINKS']["UniProt:"] for e in d['GENE']]
Note
The 2 outputs are slightly different.
- parse(res)[source]¶
Parse to any outputs returned by
KEGG.get()- Parameters
res (str) – output of a
KEGG.get().- Returns
a dictionary. Keys are those found in the KEGG entry (e.g., REACTION, ENTRY, EQUATION, …). The format of each value is various. It could be a string, a list (of strings generally), a dictionary, a float depending on the key. Depdending on the type of the entry (e.g., module, pathway), the type of the value may also differ (e.g., REACTION can be either a list of reactions or a dictionary depending on the content)
>>> # Parses a drug entry >>> res = s.get("dr:D00001") >>> # Parses a pathway entry >>> res = s.get("path:hsa10584") >>> # Parses a module entry >>> res = s.get("md:hsa_M00554") >>> # Parses a disease entry >>> res = s.get("ds:H00001") >>> # Parses a environ entry >>> res = s.get("ev:E00001") >>> # Parses Orthology entry >>> res = s.get("ko:K00001") >>> # Parses a Genome entry >>> res = s.get('genome:T00001') >>> # Parses a gene entry >>> res = s.get("hsa:1525") >>> # Parses a compound entry >>> s.get("cpd:C00001") >>> # Parses a glycan entry >>> res = s.get("gl:G00001") >>> # Parses a reaction entry >>> res = s.get("rn:R00001") >>> # Parses a rpair entry >>> res = s.get("rp:RP00001") >>> # Parses a rclass entry >>> res = s.get("rc:RC00001") >>> # Parses an enzyme entry >>> res = s.get('ec:1.1.1.1') >>> d = s.parse(res)
8.16. HGNC¶
Interface to HUGO/HGNC web services
What is HGNC ?
- URL
- Citation
“The HUGO Gene Nomenclature Committee (HGNC) has assigned unique gene symbols and names to over 37,000 human loci, of which around 19,000 are protein coding. genenames.org is a curated online repository of HGNC-approved gene nomenclature and associated resources including links to genomic, proteomic and phenotypic information, as well as dedicated gene family pages.”
—From HGNC web site, July 2013
- class HGNC(verbose=False, cache=False)[source]¶
Wrapper to the genenames web service
See details at http://www.genenames.org/help/rest-web-service-help
- fetch(database, query, frmt='json')[source]¶
Retrieve particular records from a searchable fields
Returned object is a json object with fields as in
stored_field, which is returned fromget_info()method.Only one query at a time. No wild cards are accepted.
>>> h = HGNC() >>> h.fetch('symbol', 'ZNF3') >>> h.fetch('alias_name', 'A-kinase anchor protein, 350kDa')
- get_info(frmt='json')[source]¶
Request information about the service
Fields are when the server was last updated (lastModified), the number of documents (numDoc), which fields can be queried using search and fetch (searchableFields) and which fields may be returned by fetch (storedFields).
- search(database_or_query=None, query=None, frmt='json')[source]¶
Search a searchable field (database) for a pattern
The search request is more powerful than fetch for querying the database, but search will only returns the fields hgnc_id, symbol and score. This is because this tool is mainly intended to query the server to find possible entries of interest or to check data (such as your own symbols) rather than to fetch information about the genes. If you want to retrieve all the data for a set of genes from the search result, the user could use the hgnc_id returned by search to then fire off a fetch request by hgnc_id.
- Parameters
database – if not provided, search all databases.
# Search all searchable fields for the tern BRAF h.search('BRAF') # Return all records that have symbols that start with ZNF h.search('symbol', 'ZNF*') # Return all records that have symbols that start with ZNF # followed by one and only one character (e.g. ZNF3) # Nov 2015 does not work neither here nor in within in the # official documentation h.search('symbol', 'ZNF?') # search for symbols starting with ZNF that have been approved # by HGNC h.search('symbol', 'ZNF*+AND+status:Approved') # return ZNF3 and ZNF12 h.search('symbol', 'ZNF3+OR+ZNF12') # Return all records that have symbols that start with ZNF which # are not approved (ie entry withdrawn) h.search('symbol', 'ZNF*+NOT+status:Approved')
- class HGNCDeprecated(verbose=False, cache=False)[source]¶
Interface to the HGNC service
>>> from bioservices import * >>> # Fetch XML document for gene ZAP70 >>> s = HGNC() >>> xml = s.get_xml("ZAP70") >>> # You can fetch several gene names: >>> xml = s.get_xml("ZAP70;INSR") >>> # Wrong gene name request returns an empty list >>> s.get_xml("wrong") []
For a single name, the following methods are available:
>>> # get the aliases of a given gene >>> print(s.get_aliases("ZAP70")) [u'ZAP-70', u'STD'] >>> # get UniProt accession code >>> s.get_xrefs("ZAP70")['UniProt']['xkey'] 'P43403' >>> # get XML link to a UniProt cross-reference >>> s.get_xrefs("ZAP70", "xml")['UniProt']['link'] ['http://www.uniprot.org/uniprot/P43403.xml']
You can access to the links of a cross reference as well:
values = s.get_xrefs("ZAP70") s.on_web(values['EntrezGene']['link'][0])
- References
Warning
this maybe not the official.
Constructor
- Parameters
name (str) – a name for this service
url (str) – its URL
verbose (bool) – prints informative messages if True (default is True)
requests_per_sec – maximum number of requests per seconds are restricted to 3. You can change that value. If you reach the limit, an error is raise. The reason for this limitation is that some services (e.g.., NCBI) may black list you IP. If you need or can do more (e.g., ChEMBL does not seem to have restrictions), change the value. You can also have several instance but again, if you send too many requests at the same, your future requests may be retricted. Currently implemented for REST only
All instances have an attribute called
loggingthat is an instanceof theloggingmodule. It can be used to print information, warning, error messages:self.logging.info("informative message") self.logging.warning("warning message") self.logging.error("error message")
The attribute
debugLevelcan be used to set the behaviour of the logging messages. If the argument verbose is True, the debugLebel is set to INFO. If verbose if False, the debugLevel is set to WARNING. However, you can use thedebugLevelattribute to change it to one of DEBUG, INFO, WARNING, ERROR, CRITICAL. debugLevel=WARNING means that only WARNING, ERROR and CRITICAL messages are shown.- get_xml(gene)[source]¶
Returns XML of a single gene or list of genes
- Parameters
gene (str) – a valid gene name. Several gene names can be concatenated with comma ; character (e.g., ‘ZAP70;INSR’)
>>> from bioservices import * >>> s = HGNC() >>> res = s.get_xml("ZAP70") >>> res.findAll("alias") >>> [x.text for x in res.findAll("alias")] [u'ZAP-70', u'STD']
See also
- get_xrefs(gene, keep='html')[source]¶
Get the cross references for a given single gene name
>>> databases = s.get_xrefs("ZAP70").keys() >>> # get XML link to a UniProt cross-reference >>> s.get_xrefs("ZAP70", "xml")['UniProt']['link'] ['http://www.uniprot.org/uniprot/P43403.xml']
- lookfor(pattern)[source]¶
Finds all genes that starts with a given pattern
- Parameters
pattern (str) – a string. Could be the wild character *
- Returns
list of dictionary. Each dictionary contains the ‘acc’, ‘xlink:href’ and ‘xlink:title’ keys
>>> from bioservices import * >>> s = HGNC() >>> s.lookfor("ZAP") [{'acc': 'HGNC:12858', 'xlink:href': '/HGNC/wr/gene/ZAP70', 'xlink:title': 'ZAP70'}]
This function may be used to count the number of entries:
len(s.lookfor('*'))
- mapping(value)[source]¶
maps an identifier from a database onto HGNC database
- Parameters
value (str) – a valid DB:id string (e.g. “UniProt:P36888”)
- Returns
a list of dictionary with the keys ‘acc’, ‘xlink:href’, ‘xlink:title’
>>> value = "UniProt:P43403" >>> res = s.mapping(value) >>> res[0]['xlink:title'] 'ZAP70' >>> res[0]['acc'] 'HGNC:12858'
See also
- mapping_all(entries=None)[source]¶
Retrieves cross references for more than one entry
- Parameters
entries – list of values entries (e.g., returned by the
lookfor()method.) if not provided, this method looks for all entries.- Returns
list of dictionaries with keys being all entry names. Values is a dictionary of cross references.
Warning
takes 10 minutes
8.17. Intact (complex)¶
This module provides a class IntactComplex
What is Intact Complex ?
“The Complex Portal is a manually curated, encyclopaedic resource of macromolecular complexes from a number of key model organisms.”
—From Intact web page Feb 2015
- class IntactComplex(verbose=False, cache=False)[source]¶
Interface to the Intact service
>>> from bioservices import IntactComplex >>> u = IntactComplex()
Constructor IntactComplex
- Parameters
verbose – set to False to prevent informative messages
- search(query, frmt='json', facets=None, first=None, number=None, filters=None)[source]¶
Search for a complex inside intact complex.
- Parameters
s = IntactComplex() # search for ndc80 s.search('ncd80') # Search for ndc80 and facet with the species field: s.search('ncd80', facets='species_f') # Search for ndc80 and facet with the species and biological role fields: s.search('ndc80', facets='species_f,pbiorole_f') # Search for ndc80, facet with the species and biological role # fields and filter the species using human: s.search('Ndc80', first=0, number=10, filters='species_f:("Homo sapiens")', facets='species_f,ptype_f,pbiorole_f') # Search for ndc80, facet with the species and biological role # fields and filter the species using human or mouse: s.search('Ndc80, first=0, number=10, filters='species_f:("Homo sapiens" "Mus musculus")', facets='species_f,ptype_f,pbiorole_f') # Search with a wildcard to retrieve all the information: s.search('*') # Search with a wildcard to retrieve all the information and facet # with the species, biological role and interactor type fields: s.search('*', facets='species_f,pbiorole_f,ptype_f') # Search with a wildcard to retrieve all the information, facet with # the species, biological role and interactor type fields and filter # the interactor type using small molecule: s.search('*', facets='species_f,pbiorole_f,ptype_f', filters='ptype_f:("small molecule")' # Search with a wildcard to retrieve all the information, facet with # the species, biological role and interactor type fields and filter # the interactor type using small molecule and the species using human: s.search('*', facets='species_f,pbiorole_f,ptype_f', filters='ptype_f:("small molecule"),species_f:("Homo sapiens")') # Search for GO:0016491 and paginate (first is for the offset and number # is how many do you want): s.search('GO:0016491', first=10, number=10)
The organism name used in the filter must be exact. Here is the list found by typing:
res = set(ci.search('*', frmt='pandas')['organismName'])
'Bos taurus; 9913', 'Caenorhabditis elegans; 6239', 'Canis familiaris; 9615', 'Drosophila melanogaster; 7227', 'Escherichia coli (strain K12); 83333', 'Gallus gallus; 9031', 'Homo sapiens; 9606', 'Mus musculus; 10090', 'Oryctolagus cuniculus; 9986', 'Rattus norvegicus; 10116', 'Saccharomyces cerevisiae (strain ATCC 204508 / S288c);559292', 'Schizosaccharomyces pombe (strain 972 / ATCC 24843);284812', 'Xenopus laevis; 8355'
8.18. MUSCLE¶
Interface to the MUSCLE web service
What is MUSCLE ?
“MUSCLE - (MUltiple Sequence Comparison by Log-Expectation) 1)
is claimed to achieve both better average accuracy and better speed than ClustalW or T-Coffee, depending on the chosen options. Multiple alignments of protein sequences are important in many applications, including phylogenetic tree estimation, secondary structure prediction and critical residue identification.”
—from EMBL-EBI web page
- class MUSCLE(verbose=False)[source]¶
Interface to the MUSCLE service.
>>> from bioservices import * >>> m = MUSCLE(verbose=False) >>> sequencesFasta = open('filename','r') >>> jobid = n.run(frmt="fasta", sequence=sequencesFasta.read(), email="name@provider") >>> s.getResult(jobid, "out")
Warning
It is very important to provide a real e-mail address as your job otherwise very likely will be killed and your IP, Organisation or entire domain black-listed.
Here is another similar example but we use
UniProtclass provided in bioservices to fetch the FASTA sequences:>>> from bioservices import UniProt, MUSCLE >>> u = UniProt(verbose=False) >>> f1 = u.get_fasta("P18413") >>> f2 = u.get_fasta("P18412") >>> m = MUSCLE(verbose=False) >>> jobid = m.run(frmt="fasta", sequence=f1+f2, email="name@provider") >>> m.getResult(jobid, "out")
- get_parameter_details(parameterId)[source]¶
Get detailed information about a parameter.
- Returns
An XML document providing details about the parameter or a list of values that can take the parameters if the XML could be parsed.
For example:
>>> n.get_parameter_details("format")
- get_parameters()[source]¶
List parameter names.
- Returns
An XML document containing a list of parameter names.
>>> from bioservices import muscle >>> n = muscle.Muscle() >>> res = n.get_parameters() >>> [x.text for x in res.findAll("id")]
See also
parametersto get a list of the parameters without need to process the XML output.
- get_result_types(jobid)[source]¶
Get available result types for a finished job.
- Parameters
- Returns
A dictionary, which keys correspond to the identifiers. Each identifier is itself a dictionary containing the label, description, file suffix and mediaType of the identifier.
- get_status(jobid)[source]¶
Get status of a submitted job
- Parameters
- Returns
A string giving the jobid status (e.g. FINISHED).
The values for the status are:
RUNNING: the job is currently being processed.
FINISHED: job has finished, and the results can then be retrieved.
ERROR: an error occurred attempting to get the job status.
FAILURE: the job failed.
NOT_FOUND: the job cannot be found.
- property parameters¶
- run(frmt=None, sequence=None, tree='none', email=None)[source]¶
Submit a job with the specified parameters.
Compulsary arguments
- Parameters
- Returns
A jobid that can be analysed with
getResult(),getStatus(), …
The up to data values accepted for each of these parameters can be retrieved from the
get_parameter_details().For instance,:
from bioservices import MUSCLE m = MUSCLE() m.parameterDetails("tree")
Example:
jobid = m.run(frmt="fasta", sequence=sequence_example, email="test@yahoo.fr")
frmt can be a list of formats:
frmt=['fasta','clw','clwstrict','html','msf','phyi','phys']
The returned object is a jobid, which status can be checked. It must be finished before analysing/geeting the results.
See also
getResult()
8.19. MyGeneInfo¶
Interface to the mygeneinfo web Service.
What is MyGeneInfo ?
MyGene.info provides simple-to-use REST web services to query/retrieve gene annotation data. It’s designed with simplicity and performance emphasized. You can use it to power a web application which requires querying genes and obtaining common gene annotations. For example, MyGene.info services are used to power BioGPS; or use it in an analysis pipeline to retrieve always up-to-date gene annotations.
—mygene.info home page, June 2020
- class MyGeneInfo(verbose=False, cache=False)[source]¶
Interface to mygene.infoe service
>>> from bioservices import MyGeneInfo >>> s = MyGeneInfoe()
Constructor
- Parameters
verbose (bool) – prints informative messages (default is off)
- get_genes(ids, fields='symbol,name,taxid,entrezgene,ensemblgene', species=None, dotfield=True, email=None)[source]¶
Get matching gene objects for a list of gene ids
- Parameters
ids – list of geneinfo IDs
fields (str) – a comma-separated fields to limit the fields returned from the matching gene hits. The supported field names can be found from any gene object (e.g. http://mygene.info/v3/gene/1017). Note that it supports dot notation as well, e.g., you can pass “refseq.rna”. If “fields=all”, all available fields will be returned. Default: “symbol,name,taxid,entrezgene,ensemblgene”.
species (str) – can be used to limit the gene hits from given species. You can use “common names” for nine common species (human, mouse, rat, fruitfly, nematode, zebrafish, thale-cress, frog and pig). All other species, you can provide their taxonomy ids. Multiple species can be passed using comma as a separator. Default: human,mouse,rat.
dotfield – control the format of the returned fields when passed “fields” parameter contains dot notation, e.g. “fields=refseq.rna”. If True the returned data object contains a single “refseq.rna” field, otherwise (False), a single “refseq” field with a sub-field of “rna”. Default: True.
email" (str) – If you are regular users of this services, the mygeneinfo maintainers/authors encourage you to provide an email, so that we can better track the usage or follow up with you.
mgi = MyGeneInfoe() mgi.get_genes(("301345,22637")) # first one is rat, second is mouse. This will return a 'notfound' # entry and the second entry as expected. mgi.get_genes("301345,22637", species="mouse")
- get_one_gene(geneid, fields='symbol,name,taxid,entrezgene,ensemblgene', dotfield=True, email=None)[source]¶
Get matching gene objects for one gene id
- Parameters
geneid – a valid gene ID
fields (str) – a comma-separated fields to limit the fields returned from the matching gene hits. The supported field names can be found from any gene object (e.g. http://mygene.info/v3/gene/1017). Note that it supports dot notation as well, e.g., you can pass “refseq.rna”. If “fields=all”, all available fields will be returned. Default: “symbol,name,taxid,entrezgene,ensemblgene”.
dotfield – control the format of the returned fields when passed “fields” parameter contains dot notation, e.g. “fields=refseq.rna”. If True the returned data object contains a single “refseq.rna” field, otherwise (False), a single “refseq” field with a sub-field of “rna”. Default: True.
email" (str) – If you are regular users of this services, the mygeneinfo maintainers/authors encourage you to provide an email, so that we can better track the usage or follow up with you.
mgi = MyGeneInfoe() mgi.get_genes("301345")
- get_one_query(query, email=None, dotfield=True, fields='symbol,name,taxid,entrezgene,ensemblgene', species='human,mouse,rat', size=10, _from=0, sort=None, facets=None, entrezonly=False, ensemblonly=False)[source]¶
Make gene query and return matching gene list. Support JSONP and CORS as well.
- Parameters
query (str) – Query string. Examples “CDK2”, “NM_052827”, “204639_at”, “chr1:151,073,054-151,383,976”, “hg19.chr1:151073054-151383976”. The detailed query syntax can be found from our docs.
fields (str) – a comma-separated fields to limit the fields returned from the matching gene hits. The supported field names can be found from any gene object (e.g. http://mygene.info/v3/gene/1017). Note that it supports dot notation as well, e.g., you can pass “refseq.rna”. If “fields=all”, all available fields will be returned. Default: “symbol,name,taxid,entrezgene,ensemblgene”.
species (str) – can be used to limit the gene hits from given species. You can use “common names” for nine common species (human, mouse, rat, fruitfly, nematode, zebrafish, thale-cress, frog and pig). All other species, you can provide their taxonomy ids. Multiple species can be passed using comma as a separator. Default: human,mouse,rat.
size (int) – the maximum number of matching gene hits to return (with a cap of 1000 at the moment). Default: 10.
_from (int) – the number of matching gene hits to skip, starting from 0. Combining with “size” parameter, this can be useful for paging. Default: 0.
sort – the comma-separated fields to sort on. Prefix with “-” for descending order, otherwise in ascending order. Default: sort by matching scores in decending order.
facets (str) – a single field or comma-separated fields to return facets, for example, “facets=taxid”, “facets=taxid,type_of_gene”.
entrezonly (bool) – when passed as True, the query returns only the hits with valid Entrez gene ids. Default: False.
ensembleonly (bool) – when passed as True, the query returns only the hits with valid Ensembl gene ids. Default: False.
dotfield – control the format of the returned fields when passed “fields” parameter contains dot notation, e.g. “fields=refseq.rna”. If True the returned data object contains a single “refseq.rna” field, otherwise (False), a single “refseq” field with a sub-field of “rna”. Default: True.
email" (str) – If you are regular users of this services, the mygeneinfo maintainers/authors encourage you to provide an email, so that we can better track the usage or follow up with you.
- get_queries(query, email=None, dotfield=True, scopes='all', species='human,mouse,rat', fields='symbol,name,taxid,entrezgene,ensemblgene')[source]¶
Make gene query and return matching gene list. Support JSONP and CORS as well.
- Parameters
query (str) – Query string. Examples “CDK2”, “NM_052827”, “204639_at”, “chr1:151,073,054-151,383,976”, “hg19.chr1:151073054-151383976”. The detailed query syntax can be found from our docs.
fields (str) – a comma-separated fields to limit the fields returned from the matching gene hits. The supported field names can be found from any gene object (e.g. http://mygene.info/v3/gene/1017). Note that it supports dot notation as well, e.g., you can pass “refseq.rna”. If “fields=all”, all available fields will be returned. Default: “symbol,name,taxid,entrezgene,ensemblgene”.
species (str) – can be used to limit the gene hits from given species. You can use “common names” for nine common species (human, mouse, rat, fruitfly, nematode, zebrafish, thale-cress, frog and pig). All other species, you can provide their taxonomy ids. Multiple species can be passed using comma as a separator. Default: human,mouse,rat.
dotfield – control the format of the returned fields when passed “fields” parameter contains dot notation, e.g. “fields=refseq.rna”. If True the returned data object contains a single “refseq.rna” field, otherwise (False), a single “refseq” field with a sub-field of “rna”. Default: True.
email" (str) – If you are regular users of this services, the mygeneinfo maintainers/authors encourage you to provide an email, so that we can better track the usage or follow up with you.
scopes (str) – not documented. Set to ‘all’
8.20. NCBIblast¶
Interface to the NCBIBLAST web service
What is NCBIBLAST ?
“NCBI BLAST - Protein Database Query
The emphasis of this tool is to find regions of sequence similarity, which will yield functional and evolutionary clues about the structure and function of your novel sequence.”
—from NCBIblast web page
- class NCBIblast(verbose=False)[source]¶
Interface to the NCBIblast service.
>>> from bioservices import * >>> s = NCBIblast(verbose=False) >>> jobid = s.run(program="blastp", sequence=s._sequence_example, stype="protein", database="uniprotkb", email="name@provider") >>> s.getResult(jobid, "out")
Warning
It is very important to provide a real e-mail address as your job otherwise very likely will be killed and your IP, Organisation or entire domain black-listed.
When running a blast request, a program is required. You can obtain the list using:
>>> s.parametersDetails("program") [u'blastp', u'blastx', u'blastn', u'tblastx', u'tblastn']
blastn: Search a nucleotide database using a nucleotide query
blastp: Search protein database using a protein query
blastx: Search protein database using a translated nucleotide query
tblastn Search translated nucleotide database using a protein query
tblastx Search translated nucleotide database using a translated nucleotide query
NCBIblast constructor
- Parameters
verbose (bool) – prints informative messages
- property databases¶
Returns accepted databases.
- get_parameter_details(parameterId)[source]¶
Get detailed information about a parameter.
- Returns
An XML document providing details about the parameter or a list of values that can take the parameters if the XML could be parsed.
For example:
>>> s.parameter_details("matrix") [u'BLOSUM45', u'BLOSUM50', u'BLOSUM62', u'BLOSUM80', u'BLOSUM90', u'PAM30', u'PAM70', u'PAM250']
- get_parameters()[source]¶
List parameter names.
- Returns
An XML document containing a list of parameter names.
>>> from bioservices import ncbiblast >>> n = ncbiblast.NCBIblast() >>> res = n.get_parameters() >>> [x.text for x in res.findAll("id")]
See also
parametersto get a list of the parameters without need to process the XML output.
- get_result(jobid, result_type)[source]¶
Get the job result of the specified type.
- param str jobid
a job identifier returned by
run().- param str result_type
type of result to retrieve. See
getResultTypes().
The output from the tool itself. Use the ‘format’ parameter to retireve the output in different formats, the ‘compressed’ parameter to retrieve the xml output in compressed form. Format options:
0 = pairwise, 1 = query-anchored showing identities, 2 = query-anchored no identities, 3 = flat query-anchored showing identities, 4 = flat query-anchored no identities, 5 = XML Blast output, 6 = tabular, 7 = tabular with comment lines, 8 = Text ASN.1, 9 = Binary ASN.1, 10 = Comma-separated values, 11 = BLAST archive format (ASN.1).
See NCBI Blast documentation for details. Use the ‘compressed’ parameter to return the XML output in compressed form. e.g. ‘?format=5&compressed=true’.
- get_result_types(jobid)[source]¶
Get available result types for a finished job.
- Parameters
- Returns
A dictionary, which keys correspond to the identifiers. Each identifier is itself a dictionary containing the label, description, file suffix and mediaType of the identifier.
- get_status(jobid)[source]¶
Get status of a submitted job
- Parameters
- Returns
A string giving the jobid status (e.g. FINISHED).
The values for the status are:
RUNNING: the job is currently being processed.
FINISHED: job has finished, and the results can then be retrieved.
ERROR: an error occurred attempting to get the job status.
FAILURE: the job failed.
NOT_FOUND: the job cannot be found.
- property parameters¶
- run(program=None, database=None, sequence=None, stype='protein', email=None, **kargs)[source]¶
Submit a job with the specified parameters.
Compulsary arguments
- Parameters
program (str) – BLAST program to use to perform the search (e.g., blastp)
sequence (str) – query sequence. The use of fasta formatted sequence is recommended.
database (list) – list of database names for search or possible a single string (for one database). There are some mismatch between the output of parametersDetails(“database”) and the accepted values. For instance UniProt Knowledgebase should be given as “uniprotkb”.
email (str) – a valid email address. Will be checked by the service itself.
Optional arguments. If not provided, a default value will be used
- Parameters
type (str) – query sequence type in ‘dna’, ‘rna’ or ‘protein’ (default is protein).
matrix (str) – scoring matrix to be used in the search (e.g., BLOSUM45).
gapalign (bool) – perform gapped alignments.
alignments (int) – maximum number of alignments displayed in the output.
exp – E-value threshold.
filter (bool) – low complexity sequence filter to process the query sequence before performing the search.
scores (int) – maximum number of scores displayed in the output.
dropoff (int) – amount score must drop before extension of hits is halted.
match_scores – match/miss-match scores to generate a scoring matrix for nucleotide searches.
gapopen (int) – penalty for the initiation of a gap.
gapext (int) – penalty for each base/residue in a gap.
seqrange – region of the query sequence to use for the search. Default: whole sequence.
- Returns
A jobid that can be analysed with
getResult(),getStatus(), …
The up to data values accepted for each of these parameters can be retrieved from the
get_parameter_details().For instance,:
from bioservices import NCBIblast n = NCBIblast() n.get_parameter_details("program")
Example:
jobid = n.run(program="blastp", sequence=n._sequence_example, stype="protein", database="uniprotkb", email="test@yahoo.fr")
Database can be a list of databases:
database=["uniprotkb", "uniprotkb_swissprot"]
The returned object is a jobid, which status can be checked. It must be finished before analysing/geeting the results.
See also
getResult()Warning
Cases are not important. Spaces in the database case should be replaced by underscore.
Note
database returned by the server have meaningless names since they do not map to the expected names. An example is “ENA Sequence Release” that should be provided as em_rel
http://www.ebi.ac.uk/Tools/sss/ncbiblast/help/index-nucleotide.html
8.21. OmniPath Commons¶
Interface to OmniPath web service
What is OmniPath ?
A comprehensive collection of literature curated human signaling pathways.
—From OmniPath web site, March 2016
- class OmniPath(verbose=False, cache=False)[source]¶
Interface to the OmniPath service
>>> from bioservices import OmniPath >>> o = OmniPath() >>> net = o.get_network() >>> interactions = o.get_interactions('P00533')
Constructor OmniPath
- Parameters
verbose – set to False to prevent informative messages
- get_interactions(query='', frmt='json', fields=[])[source]¶
Interactions of proteins
- Parameters
Example:
res_one = o.get_interactions('P00533') res_many = o.get_interactions('P00533,O15117,Q96FE5') res_many = o.get_interactions(['P00533','O15117','Q96FE5']) res_one = o.get_interactions('P00533', fields='sources') res_one = o.get_interactions('P00533', fields=['source']) res_one = o.get_interactions('P00533', fields=['source', 'references'])
You may also keep query to an empty string, but the entire DB will then be downloaded. This may take time and the
timeoutmay need to be increased manually.If frmt is set to TSV, the output is a TSV table with a header. If set to json, a dictionary is returned.
- get_ptms(query='', ptm_type=None, frmt='json', fields=[])[source]¶
List enzymes, substrates and PTMs
- Parameters
query (str) – a valid uniprot identifier (e.g. P00533). It can also be a list of uniprot identifiers, or a string with comma-separated identifiers.
ptm_type (str) – restrict the output to this type of PTM (e.g., phosphorylation)
fields (str) – additional fields to be added to the output (e.g., sources, references)
8.22. Panther¶
Interface to some part of the Panther web service
What is Panther ?
- URL
- Citation
The PANTHER (Protein ANalysis THrough Evolutionary Relationships) Classification System was designed to classify proteins (and their genes) in order to facilitate high-throughput analysis. Proteins have been classified according to:
Family and subfamily: families are groups of evolutionarily related proteins; subfamilies are related proteins that also have the same function
Molecular function: the function of the protein by itself or with directly interacting proteins at a biochemical level, e.g. a protein kinase
Biological process: the function of the protein in the context of a larger network of proteins that interact to accomplish a process at the level of the cell or organism, e.g. mitosis.
Pathway: similar to biological process, but a pathway also explicitly specifies the relationships between the interacting molecules.
—From PantherDB (about) , Feb 2020
- class Panther(verbose=True, cache=False)[source]¶
Interface to Panther pages
>>> from bioservics import Panther >>> p = Panther() >>> p.get_supported_genomes() >>> p.get_ortholog("zap70", 9606) >>> from bioservics import Panther >>> p = Panther() >>> taxon = [x[0]['taxon_id'] for x in p.get_supported_genomes() if "coli" in x['name'].lower()] >>> # you may also use our method called search_organism >>> taxon = p.get_taxon_id(pattern="coli") >>> res = p.get_mapping("abrB,ackA,acuI", taxon)
The get_mapping returns for each gene ID the GO terms corresponding to each ID. Those go terms may belong to different categories (see meth:get_annotation_datasets):
MF for molecular function
BP for biological process
PC for Protein class
CC Cellular location
Pathway
Note that results from the website application http://pantherdb.org/ do not agree with the oupput of the get_mapping service… Try out the dgt gene from ecoli for example
Constructor
- Parameters
verbose – set to False to prevent informative messages
- get_enrichment(gene_list, organism, annotation, enrichment_test='Fisher', correction='FDR', ref_gene_list=None)[source]¶
Returns over represented genes
Compares a test gene list to a reference gene list, and determines whether a particular class (e.g. molecular function, biological process, cellular component, PANTHER protein class, the PANTHER pathway or Reactome pathway) of genes is overrepresented or underrepresented.
- Parameters
organism – a valid taxon ID
enrichment_test – either Fisher or Binomial test
correction – correction for multiple testing. Either FDR, Bonferonni, or None.
annotation – one of the supported PANTHER annotation data types. See
get_annotation_datasets()to retrieve a list of supported annotation data typesref_gene_list – if not specified, the system will use all the genes for the specified organism. Otherwise, a list delimited by comma. Maximum of 100000 Identifiers can be any of the following: Ensemble gene identifier, Ensemble protein identifier, Ensemble transcript identifier, Entrez gene id, gene symbol, NCBI GI, HGNC Id, International protein index id, NCBI UniGene id, UniProt accession andUniProt id.
- Returns
a dictionary with the following keys. ‘reference’ contains the orgnaism, ‘input_list’ is the input gene list with unmapped genes. ‘result’ contains the list of candidates.
>>> from bioservices import Panther >>> p = Panther() >>> res = p.get_enrichment('zap70,mek1,erk', 9606, "GO:0008150") >>> For molecular function, use : >>> res = p.get_enrichment('zap70,mek1,erk', 9606, "ANNOT_TYPE_ID_PANTHER_GO_SLIM_MF")
- get_family_msa(family, taxon_list=None)[source]¶
Returns MSA information for the specified family.
- Parameters
family – family ID
taxon_list – Zero or more taxon IDs separated by ‘,’.
- get_family_ortholog(family, taxon_list=None)[source]¶
Search for matching orthologs in target organisms
Also return the corresponding position in the target organism sequence. The system searches for matching orthologs in the gene family that contains the search gene associated with the search term.
- Parameters
family – Family ID
taxon_list – Zero or more taxon IDs separated by ‘,’.
- get_homolog_position(gene, organism, position, ortholog_type='all')[source]¶
- Parameters
gene – Can be any of the following: Ensemble gene identifier, Ensemble protein identifier, Ensemble transcript identifier, Entrez gene id, gene symbol, NCBI GI, HGNC Id, International protein index id, NCBI UniGene id, UniProt accession andUniProt id
organism – a valid taxon ID
ortholog_type – optional parameter to specify ortholog type of target organism
- get_mapping(gene_list, taxon)[source]¶
Map identifiers
Each identifier to be delimited by comma i.e. ‘,. Maximum of 1000 Identifiers can be any of the following: Ensemble gene identifier, Ensemble protein identifier, Ensemble transcript identifier, Entrez gene id, gene symbol, NCBI GI, HGNC Id, International protein index id, NCBI UniGene id, UniProt accession and UniProt id
- Parameters
gene_list – see above
taxon – one taxon ID. See supported
get_supported_genomes()
If an identifier is not found, information can be found in the unmapped_genes key while found identifiers are in the mapped_genes key.
Warning
found and not found identifiers are dispatched into unmapped and mapped genes. If there are not found identifiers, the input gene list and the mapped genes list do not have the same length. The input names are not stored in the output. Developpers should be aware of that feature.
- get_ortholog(gene_list, organism, target_organism=None, ortholog_type='all')[source]¶
search for matching orthologs in target organisms.
Searches for matching orthologs in the gene family that contains the search gene associated with the search terms. Returns ortholog genes in target organisms given a search organism, the search terms and a list of target organisms.
- Parameters
gene_list –
organism – a valid taxon ID
target_organism – zero or more taxon IDs separated by ‘,’. See
get_supported_genomes()ortholog_type – optional parameter to specify ortholog type of target organism
- Returns
a dictionary with “mapped” and “unmapped” keys, each of them being a list. For each unmapped gene, a dictionary with id and organism is is returned. For the mapped gene, a list of ortholog is returned.
- get_supported_families(N=1000, progress=True)[source]¶
Returns the list of supported PANTHER family IDs
This services returns only 1000 items per request. This is defined by the index. For instance index set to 1 returns the first 1000 families. Index set to 2 returns families between index 1000 and 2000 and so on. As of 20 Feb 2020, there was about 15,000 families.
This function simplifies your life by calling the service as many times as required. Therefore it returns all families in one go.
- get_supported_genomes(type=None)[source]¶
Returns list of supported organisms.
- Parameters
type – can be chrLoc to restrict the search
8.23. Pathway Commons¶
This module provides a class PathwayCommons
What is PathwayCommons ?
- URL
- REST
Pathway Commons is a convenient point of access to biological pathway information collected from public pathway databases, which you can search, visualize and download. All data is freely available, under the license terms of each contributing database.
—PathwayCommons home page, Nov 2013
Data is freely available, under the license terms of each contributing database.
- class PathwayCommons(verbose=True, cache=False)[source]¶
Interface to the PathwayCommons service
>>> from bioservices import * >>> pc2 = PathwayCommons(verbose=False) >>> res = pc2.get("http://identifiers.org/uniprot/Q06609")
Todo
traverse() method not implemented.
Constructor
- Parameters
verbose (bool) – prints informative messages
- property default_extension¶
set extension of the requests (default is json). Can be ‘json’ or ‘xml’
- get(uri, frmt='BIOPAX')[source]¶
Retrieves full pathway information for a set of elements
elements can be for example pathway, interaction or physical entity given the RDF IDs. Get commands only retrieve the BioPAX elements that are directly mapped to the ID. Use the
traverse()query to traverse BioPAX graph and obtain child/owner elements.- Parameters
uri (str) – valid/existing BioPAX element’s URI (RDF ID; for utility classes that were “normalized”, such as entity refereneces and controlled vocabularies, it is usually a Identifiers.org URL. Multiple IDs can be provided using list uri=[http://identifiers.org/uniprot/Q06609, http://identifiers.org/uniprot/Q549Z0’] See also about MIRIAM and Identifiers.org.
format (str) – output format (values)
- Returns
a complete BioPAX representation for the record pointed to by the given URI is returned. Other output formats are produced by converting the BioPAX record on demand and can be specified by the optional format parameter. Please be advised that with some output formats it might return “no result found” error if the conversion is not applicable for the BioPAX result. For example, BINARY_SIF output usually works if there are some interactions, complexes, or pathways in the retrieved set and not only physical entities.
>>> from bioservices import PathwayCommons >>> pc2 = PathwayCommons(verbose=False) >>> res = pc2.get("col5a1") >>> res = pc2.get("http://identifiers.org/uniprot/Q06609")
- get_sifgraph_common_stream(source, limit=1, direction='DOWNSTREAM', pattern=None)[source]¶
finds the common stream for them; extracts a sub-network from the loaded Pathway Commons SIF model.
- Parameters
source – set of gene identifiers (HGNC symbol). Can be a list of identifiers or just one string(if only one identifier)
limit (int) – Graph traversal depth. Limit > 1 value can result in very large data or error.
direction (str) – Graph traversal direction. Use UNDIRECTED if you want to see interacts-with relationships too.
pattern (str) – Filter by binary relationship (SIF edge) type(s). one of “BOTHSTREAM”, “UPSTREAM”, “DOWNSTREAM”, “UNDIRECTED”.
- returns: the graph in SIF format. The output must be stripped and
returns one line per relation. In each line, items are separated by a tabulation. You can save the text with .sif extensions and it should be ready to use e.g. in cytoscape viewer.
res = pc.get_sifgraph_common_stream(['BRD4', 'MYC'])
- get_sifgraph_neighborhood(source, limit=1, direction='BOTHSTREAM', pattern=None)[source]¶
finds the neighborhood sub-network in the Pathway Commons Simple Interaction Format (extented SIF) graph (see http://www.pathwaycommons.org/pc2/formats#sif)
- Parameters
source – set of gene identifiers (HGNC symbol). Can be a list of identifiers or just one string(if only one identifier)
limit (int) – Graph traversal depth. Limit > 1 value can result in very large data or error.
direction (str) – Graph traversal direction. Use UNDIRECTED if you want to see interacts-with relationships too.
pattern (str) – Filter by binary relationship (SIF edge) type(s). one of “BOTHSTREAM”, “UPSTREAM”, “DOWNSTREAM”, “UNDIRECTED”.
- returns: the graph in SIF format. The output must be stripped and
returns one line per relation. In each line, items are separated by a tabulation. You can save the text with .sif extensions and it should be ready to use e.g. in cytoscape viewer.
res = pc.get_sifgraph_neighborhood('BRD4')
- get_sifgraph_pathsbetween(source, limit=1, directed=False, pattern=None)[source]¶
finds the paths between them; extracts a sub-network from the Pathway Commons SIF graph.
- Parameters
source – set of gene identifiers (HGNC symbol). Can be a list of identifiers or just one string(if only one identifier)
limit (int) – Graph traversal depth. Limit > 1 value can result in very large data or error.
directed (bool) – Directionality: ‘true’ is for DOWNSTREAM/UPSTREAM, ‘false’ - UNDIRECTED
pattern (str) – Filter by binary relationship (SIF edge) type(s). one of “BOTHSTREAM”, “UPSTREAM”, “DOWNSTREAM”, “UNDIRECTED”.
- returns: the graph in SIF format. The output must be stripped and
returns one line per relation. In each line, items are separated by a tabulation. You can save the text with .sif extensions and it should be ready to use e.g. in cytoscape viewer.
- get_sifgraph_pathsfromto(source, target, limit=1, pattern=None)[source]¶
finds the paths between them; extracts a sub-network from the Pathway Commons SIF graph.
- Parameters
source – set of gene identifiers (HGNC symbol). Can be a list of identifiers or just one string(if only one identifier)
param target: A target set of gene identifiers. :param int limit: Graph traversal depth. Limit > 1 value can result
in very large data or error.
- Parameters
pattern (str) – Filter by binary relationship (SIF edge) type(s). one of “BOTHSTREAM”, “UPSTREAM”, “DOWNSTREAM”, “UNDIRECTED”.
- returns: the graph in SIF format. The output must be stripped and
returns one line per relation. In each line, items are separated by a tabulation. You can save the text with .sif extensions and it should be ready to use e.g. in cytoscape viewer.
- graph(kind, source, target=None, direction=None, limit=1, frmt=None, datasource=None, organism=None)[source]¶
Finds connections and neighborhoods of elements
Connections can be for example the shortest path between two proteins or the neighborhood for a particular protein state or all states.
Graph searches take detailed BioPAX semantics such as generics or nested complexes into account and traverse the graph accordingly. The starting points can be either physical entites or entity references.
In the case of the latter the graph search starts from ALL the physical entities that belong to that particular entity references, i.e. all of its states. Note that we integrate BioPAX data from multiple databases based on our proteins and small molecules data warehouse and consistently normalize UnificationXref, EntityReference, Provenance, BioSource, and ControlledVocabulary objects when we are absolutely sure that two objects of the same type are equivalent. We, however, do not merge physical entities and reactions from different sources as matching and aligning pathways at that level is still an open research problem. As a result, graph searches can return several similar but disconnected sub-networks that correspond to the pathway data from different providers (though some physical entities often refer to the same small molecule or protein reference or controlled vocabulary).
- Parameters
kind (str) – graph query
source (str) – source object’s URI/ID. Multiple source URIs/IDs must be encoded as list of valid URI source=[‘http://identifiers.org/uniprot/Q06609’, ‘http://identifiers.org/uniprot/Q549Z0’].
target (str) – required for PATHSFROMTO graph query. target URI/ID. Multiple target URIs must be encoded as list (see source parameter).
direction (str) – graph search direction in [BOTHSTREAM, DOWNSTREAM, UPSTREAM] see
_valid_directionsattribute.limit (int) – graph query search distance limit (default = 1).
format (str) – output format. see
_valid-formatdatasource (str) – datasource filter (same as for ‘search’).
organism (str) – organism filter (same as for ‘search’).
- Returns
By default, graph queries return a complete BioPAX representation of the subnetwork matched by the algorithm. Other output formats are available as specified by the optional format parameter. Please be advised that some output format choices might cause “no result found” error if the conversion is not applicable for the BioPAX result (e.g., BINARY_SIF output fails if there are no interactions, complexes, nor pathways in the retrieved set).
>>> from bioservices import PathwayCommons >>> pc2 = PathwayCommons(verbose=False) >>> res = pc2.graph(source="http://identifiers.org/uniprot/P20908", kind="neighborhood", format="EXTENDED_BINARY_SIF")
- search(q, page=0, datasource=None, organism=None, type=None)[source]¶
Text search in PathwayCommons using Lucene query syntax
Some of the parameters are BioPAX properties, others are composite relationships.
All index fields are (case-sensitive): comment, ecnumber, keyword, name, pathway, term, xrefdb, xrefid, dataSource, and organism.
The pathway field maps to all participants of pathways that contain the keyword(s) in any of its text fields.
Finally, keyword is a transitive aggregate field that includes all searchable keywords of that element and its child elements.
All searches can also be filtered by data source and organism.
It is also possible to restrict the domain class using the ‘type’ parameter.
This query can be used standalone or to retrieve starting points for graph searches.
- Parameters
q (str) – requires a keyword , name, external identifier, or a Lucene query string.
page (int) – (N>=0, default is 0), search result page number.
datasource (str) – filter by data source (use names or URIs of pathway data sources or of any existing Provenance object). If multiple data source values are specified, a union of hits from specified sources is returned. datasource=[reactome,pid] returns hits associated with Reactome or PID.
organism (str) – The organism can be specified either by official name, e.g. “homo sapiens” or by NCBI taxonomy id, e.g. “9606”. Similar to data sources, if multiple organisms are declared a union of all hits from specified organisms is returned. For example organism=[9606, 10016] returns results for both human and mice.
type (str) – BioPAX class filter. (e.g., ‘pathway’, ‘proteinreference’)
>>> from bioservices import PathwayCommons >>> pc2 = PathwayCommons(vverbose=False) >>> pc2.search("Q06609") >>> pc2.search("brca2", type="proteinreference", organism="homo sapiens", datasource="pid") >>> pc2.search("name:'col5a1'", type="proteinreference", organism=9606) >>> pc2.search("a*", page=3)
Find the FGFR2 keyword:
pc2.search("FGFR2")
Find pathways by FGFR2 keyword in any index field.:
pc2.search("FGFR2", type="pathway")
Finds control interactions that contain the word binding but not transcription in their indexed fields:
pc2.search("binding NOT transcription", type="control")
Find all interactions that directly or indirectly participate in a pathway that has a keyword match for “immune” (Note the star after immune):
pc.search(“pathway:immune*”, type=”conversion”)
Find all Reactome pathways:
pc.search("*", type="pathway", datasource="reactome")
- top_pathways(query='*', datasource=None, organism=None)[source]¶
This command returns all top pathways
Pathways can be top or pathways that are neither ‘controlled’ nor ‘pathwayComponent’ of another process.
- param query
a keyword, name, external identifier or lucene query string like in ‘search’. Default is “*”
- param str datasource
filter by data source (same as search)
- param str organism
organism filter. 9606 for human.
- return
dictionary with information about top pathways. Check the “searchHit” key for information about “dataSource” for instance
>>> from bioservices import PathwayCommons >>> pc2 = PathwayCommons(verbose=False) >>> res = pc2.top_pathways()
- traverse(uri, path)[source]¶
Provides XPath-like access to the PC.
The format of the path query is in the form:
[InitialClass]/[property1]:[classRestriction(optional)]/[property2]... A "*"
sign after the property instructs path accessor to transitively traverse that property. For example, the following path accessor will traverse through all physical entity components within a complex:
"Complex/component*/entityReference/xref:UnificationXref"The following will list display names of all participants of interactions, which are components (pathwayComponent) of a pathway (note: pathwayOrder property, where same or other interactions can be reached, is not considered here):
"Pathway/pathwayComponent:Interaction/participant*/displayName"The optional parameter classRestriction allows to restrict/filter the returned property values to a certain subclass of the range of that property. In the first example above, this is used to get only the Unification Xrefs. Path accessors can use all the official BioPAX properties as well as additional derived classes and parameters in paxtools such as inverse parameters and interfaces that represent anonymous union classes in OWL. (See Paxtools documentation for more details).
- Parameters
See also
- Returns
XML result that follows the Search Response XML Schema (TraverseResponse type; pagination is disabled: returns all values at once)
from bioservices import PathwayCommons pc2 = PathwayCommons(verbose=False) res = pc2.traverse(uri=['http://identifiers.org/uniprot/P38398','http://identifiers.org/uniprot/Q06609'], path="ProteinReference/organism") res = pc2.traverse(uri="http://identifiers.org/uniprot/Q06609", path="ProteinReference/entityReferenceOf:Protein/name") res = pc2.traverse("http://identifiers.org/uniprot/P38398", path="ProteinReference/entityReferenceOf:Protein") res = pc2.traverse(uri=["http://identifiers.org/uniprot/P38398", "http://identifiers.org/taxonomy/9606"], path="Named/name")
8.24. PDB/PDBe modules¶
Interface to the PDB web Service (New API Jan 2021).
What is PDB ?
An Information Portal to Biological Macromolecular Structures
—PDB home page, Jan 2021
- class PDB(verbose=False, cache=False)[source]¶
Interface to PDB service (new API Jan 2021)
With the new API, one method called
search()is provided by PDB. To perform a search you need to define a query. Here is an example>>> from bioservices import PDB >>> s = PDB() >>> query = {"query": ... {"type": "terminal", ... "service": "text", ... "parameters": { ... "value": "thymidine kinase" ... } ... }, ... "return_type": "entry"} >>> res = s.search(query, return_type=return_type)
Note
as of December 2020, a new API has be set up by PDB. some prevous functionalities such as return list of Ligand are not supported anymore (Jan 2021). However, many more powerful searches as available. I encourage everyone to look at the PDB page for complex examples: http://search.rcsb.org/#examples
As mentionnaed above, the PDB service provide one method called search available in
search(). We will not cover all the power and capability of this search function. User should refer to the official PDB help for that. Yet, given examples from PDB should all work with this method.When possible, we will add convenient aliases function in this class. For now we have for example the
get_current_ids()andget_similarity_sequence()that users may find useful.The main idea behind the PDB API is to create queries that can access to different type of services. A query will need to at least two keys:
query
return_type
Consider this basic example that searches for the text thymidine kinase:
{ "query": { "type": "terminal", "service": "text", "parameters": { "value": "thymidine kinase" } }, "return_type": "entry" }
Here the query is defined by a query and a return_type indeed. The return type is a simple value such as entry. The query itself is composed of 3 pairs of key/value. Here we have the type service and parameters as defined below.
The query can have several fields:
type: the clause type can be either terminal or group
terminal: performs an atomic search operation, e.g. searches for a particular value in a particular field.
group: wraps other terminal or group nodes and is used to combine multiple queries in a logical fashion.
service:
text: linguistic searches against textual annotations.
sequence: uses MMSeq2 to perform sequence matching searches (blast-like). following targets that are available:
pdb_protein_sequence,
pdb_dna_sequence,
pdb_na_sequence
seqmotif: performs short motif searches against nucleotide or protein sequences using 3 different inputs:
simple (e.g., CXCXXL)
prosite (e.g., C-X-C-X(2)-[LIVMYFWC])
regex (e.g., CXCX{2}[LIVMYFWC])
structure: searches matching a global 3D shape of assemblies or chains of a given entry (identified by PDB ID), in either strict (strict_shape_match) or relaxed (relaxed_shape_match) modes
strucmotif: Performs structural motif searches on all available PDB structures.
chemical: queries of small-molecule constituents of PDB structures, based on chemical formula and chemical structure. Queries for matching and similar chemical structures can be performed using SMILES and InChI descriptors as search targets.
graph-strict: atom type, formal charge, bond order, atom and bond chirality, aromatic assignment are used as matching criteria for this search type.
graph-relaxed: atom type, formal charge and bond order are used as matching criteria for this search type.
graph-relaxed-stereo: atom type, formal charge, bond order, atom and bond chirality are used as matching criteria for this search type.
fingerprint-similarity: Tanimoto similarity is used as the matching criteria
Concerning the return_type key, it can be one of :
entry: a list of PDB IDs.
assembly: list of PDB IDs appended with assembly IDs in the format of a [pdb_id]-[assembly_id], corresponding to biological assemblies.
polymer_entity: list of PDB IDs appended with entity IDs in the format of a [pdb_id]_[entity_id], corresponding to polymeric molecular entities.
non_polymer_entity: list of PDB IDs appended with entity IDs in the format of a [pdb_id]_[entity_id], corresponding to non-polymeric entities (or ligands).
polymer_instance: list of PDB IDs appended with asym IDs in the format of a [pdb_id].[asym_id], corresponding to instances of certain polymeric molecular entities, also known as chains.
Optional arguments
There are many optional arguments. Let us see a couple of them. Pagination can be set (default is 10 entries) using the request_options (optional) key. Consider this query example:
{ "query": { "type": "terminal", "service": "text", "parameters": { "attribute": "rcsb_polymer_entity.formula_weight", "operator": "greater", "value": 500 } }, "request_options": { "pager": { "start": 0, "rows": 100 } }, "return_type": "polymer_entity" }
Here, the query searches for the polymer_entity that have a formula weight above 500. Withe request_options pager set to 100, we will get the first 100 hits.
To return all hits, set this field in the request_options:
"return_all_hits": true
Coming back at the first basic example, we can reuse it to illustrate how to refine the search using attribute and operators:
{ "query": { "type": "terminal", "service": "text", "parameters": { "value": "thymidine kinase", "attribute": "exptl.method", "operator": "exact_match", } }, "return_type": "entry" }
All valid combo of operators and attributes can be found here: http://search.rcsb.org/search-attributes.html
For instance, in the example above only in, exact_match and exists can be used with exptl.method attribute. This is not checked in bioservices.
Sorting is determined by the sort object in the request_options context. It allows you to add one or more sorting conditions to control the order of the search result hits. The sort operation is defined on a per field level, with special field name for score to sort by score (the default)<
By default sorting is done in descending order (“desc”). The sort can be reversed by setting direction property to “asc”. This example demonstrates how to sort the search results by release date:
{ "query": { "type": "terminal", "service": "text", "parameters": { "attribute": "struct.title", "operator": "contains_phrase", "value": ""hiv protease"" } }, "request_options": { "sort": [ { "sort_by": "rcsb_accession_info.initial_release_date", "direction": "desc" } ] }, "return_type": "entry" }
Again, many more complex examples can be found on PDB page.
Constructor
- Parameters
verbose (bool) – prints informative messages (default is off)
- get_similarity_sequence(seq)[source]¶
Search of seauence similarity search with protein sequence
seq = “VLSPADKTNVKAAWGKVGAHAGEYGAEALERMFLSFPTTKTYFPHFDLSHGSAQVKGHGKKVADALTAVAHVDDMPNAL” results = p.get_similarity_sequence(seq)
- search(query, request_options=None, request_info=None, return_type=None)[source]¶
search request represented as a JSON object.
This is the only function in PDB API. You should be able to perform any valid PDB searches here (see the
bioservices.pdb.PDBdocumentation for details. Note, however, that we have aliases methods in BioServices that will be added on demand for common searches.- Parameters
query (str) – the search expression. Can be omitted if, instead of IDs retrieval, facets or count operation should be performed. In this case the request must be configured via the request_options context.
request_options (str) – (optional) controls various aspects of the search request including pagination, sorting, scoring and faceting.
request_info (str) – additional information about the query, e.g. query_id. (optional)
return_type (str) – type of results to return.
- Returns
json results
You must define a query as defined in the PDB web page. For example the following query search for macromolecular PDB entities that share 90% sequence identity with GTPase HRas protein from Gallus gallus (Chicken):
query = { "query": { "type": "terminal", "service": "sequence", "parameters": { "evalue_cutoff": 1, "identity_cutoff": 0.9, "target": "pdb_protein_sequence", "value": "MTEYKLVVVGAGGVGKSALTIQLIQNHFVDEYDPTIEDSYRKQVVIDGETCLLDILDTAGQEEYSAMRDQYMRTGEGFLCVFAINNTKSFEDIHQYREQIKRVKDSDDVPMVLVGNKCDLPARTVETRQAQDLARSYGIPYIETSAKTRQGVEDAFYTLVREIRQHKLRKLNPPDESGPGCMNCKCVIS" } }, "request_options": { "scoring_strategy": "sequence" }, "return_type": "polymer_entity" }
What is important is that the dictionary called query contains 2 compulsary keys namely query and return_type. The two other optional keys are request_options and return_info
You would then call the PDB search as follows:
from bioservices import PDB p = PDB() results = p.search(query)
Now, in BioServices, you can also decompose the query as follows:
query = { "type": "terminal", "service": "sequence", "parameters": { "evalue_cutoff": 1, "identity_cutoff": 0.9, "target": "pdb_protein_sequence", "value": "MTEYKLVVVGAGGVGKSALTIQLIQNHFVDEYDPTIEDSYRKQVVIDGETCLLDILDTAGQEEYSAMRDQYMRTGEGFLCVFAINNTKSFEDIHQYREQIKRVKDSDDVPMVLVGNKCDLPARTVETRQAQDLARSYGIPYIETSAKTRQGVEDAFYTLVREIRQHKLRKLNPPDESGPGCMNCKCVIS" }} request_options = { "scoring_strategy": "sequence"} return_type= "polymer_entity"
and then use PDB search again:
from bioservices import PDB p = PDB() results = p.search(query, request_options=request_options, return_type=return_type)
or even simpler for the Pythonic lovers:
results = p.search(**query)
Interface to the PDBe web Service.
What is PDBe ?
PDBe is a founding member of the Worldwide Protein Data Bank which collects, organises and disseminates data on biological macromolecular structures. In collaboration with the other Worldwide Protein Data Bank (wwPDB) partners, we work to collate, maintain and provide access to the global repository of macromolecular structure models, the Protein Data Bank (PDB).
—PDBe home page, June 2020
- class PDBe(verbose=False, cache=False)[source]¶
Interface to part of the PDBe service
>>> from bioservices import PDBe >>> s = PDBe() >>> res = s.get_file("1FBV", "pdb")
Constructor
- Parameters
verbose (bool) – prints informative messages (default is off)
- get_assembly(query)[source]¶
Provides information for each assembly of a given PDB ID. T
This information is broken down at the entity level for each assembly. The information given includes the molecule name, type and class, the chains where the molecule occur, and the number of copies of each entity in the assembly.
- Parameters
query – a 4-character PDB id code
p.get_assembly('1cbs')
- get_binding_sites(query)[source]¶
Pprovides details on binding sites in the entry
STRUCT_SITE records in PDB files (or mmcif equivalent thereof), such as ligand, residues in the site, description of the site, etc.
- Parameters
query – a 4-character PDB id code
p.get_binding_sites('1cbs')
- get_drugbank_annotation(query)[source]¶
This call provides DrugBank annotation of all ligands, i.e. ‘bound’
- Parameters
query – a 4-character PDB id code
p.get_drugbank_annotation('5hht')
- get_electron_density_statistics(query)[source]¶
This call details the statistics for electron density.
- Parameters
query – a 4-character PDB id code
p.get_electron_density_statistics('1cbs')
- get_experiment(query)[source]¶
Provides details of experiment(s) carried out in determining the structure of the entry.
Each experiment is described in a separate dictionary. For X-ray diffraction, the description consists of resolution, spacegroup, cell dimensions, R and Rfree, refinement program, etc. For NMR, details of spectrometer, sample, spectra, refinement, etc. are included. For EM, details of specimen, imaging, acquisition, reconstruction, fitting etc. are included.
- Parameters
query – a 4-character PDB id code
p.get_experiment('1cbs')
- get_files(query)[source]¶
Provides URLs and brief descriptions (labels) for PDB entry
Also, for mmcif files, biological assembly files, FASTA file for sequences, SIFTS cross reference XML files, validation XML files, X-ray structure factor file, NMR experimental constraints files, etc.
- Parameters
query – a 4-character PDB id code
p.get_files('1cbs')
- get_functional_annotation(query)[source]¶
Provides functional annotation of all ligands, i.e. ‘bound’
- Parameters
query – a 4-character PDB id code
p.get_functional_annotation('1cbs')
- get_ligand_monomers(query)[source]¶
Provides a a list of modelled instances of ligands,
ligands i.e. ‘bound’ molecules that are not waters.
- Parameters
query – a 4-character PDB id code
p.get_ligand_monomers('1cbs')
- get_modified_residues(query)[source]¶
Provides a list of modelled instances of modified amino acids or nucleotides in protein, DNA or RNA chains.
- Parameters
query – a 4-character PDB id code
p.get_modified_residues('4v5j')
- get_molecules(query)[source]¶
Return details of molecules (or entities in mmcif-speak) modelled in the entry
This can be entity id, description, type, polymer-type (if applicable), number of copies in the entry, sample preparation method, source organism(s) (if applicable), etc.
- Parameters
query – a 4-character PDB id code
p.get_molecules('1cbs')
- get_mutated_residues(query)[source]¶
Provides a list of modelled instances of mutated amino acids or nucleotides in protein, DNA or RNA chains.
- Parameters
query – a 4-character PDB id code
p.get_mutated_residues('1bgj')
- get_nmr_resources(query)[source]¶
This call provides URLs of available additional resources for NMR entries. E.g., mapping between structure (PDB) and chemical shift (BMRB) entries. :param query: a 4-character PDB id code
p.get_nmr_resources('1cbs')
- get_observed_ranges(query)[source]¶
- Provides observed ranges, i.e., segments of structural coverage of
polymeric molecues that are modelled fully or partly
- Parameters
query – a 4-character PDB id code
p.get_observed_ranges('1cbs')
- get_observed_ranges_in_pdb_chain(query, chain_id)[source]¶
- Provides observed ranges, i.e., segments of structural coverage of
polymeric molecules in a particular chain
- Parameters
query – a 4-character PDB id code
query – a PDB chain ID
p.get_observed_ranges_in_pdb_chain('1cbs', "A")
- get_observed_residues_ratio(query)[source]¶
Provides the ratio of observed residues for each chain in each molecule
- The list of chains within an entity is sorted by observed_ratio (descending order),
partial_ratio (ascending order), and number_residues (descending order).
- Parameters
query – a 4-character PDB id code
p.get_observed_residues_ratio('1cbs')
Provides DOI’s for related raw experimental datasets
Includes diffraction image data, small-angle scattering data and electron micrographs.
- Parameters
query – a 4-character PDB id code
p.get_cofactor('5o8b')
Return publications obtained from both EuroPMC and UniProt. T
These are articles which cite the primary citation of the entry, or open-access articles which mention the entry id without explicitly citing the primary citation of an entry.
- Parameters
query – a 4-character PDB id code
p.get_related_publications('1cbs')
- get_release_status(query)[source]¶
Provides status of a PDB entry (released, obsoleted, on-hold etc) along with some other information such as authors, title, experimental method, etc.
- Parameters
query – a 4-character PDB id code
p.get_release_status('1cbs')
- get_residue_listing(query)[source]¶
Provides lists all residues (modelled or otherwise) in the entry.
Except waters, along with details of the fraction of expected atoms modelled for the residue and any alternate conformers.
- Parameters
query – a 4-character PDB id code
p.get_residue_listing('1cbs')
- get_residue_listing_in_pdb_chain(query, chain_id)[source]¶
Provides all residues (modelled or otherwise) in the entry
Except waters, along with details of the fraction of expected atoms modelled for the residue and any alternate conformers.
- Parameters
query – a 4-character PDB id code
query – a PDB chain ID
p.get_residue_listing_in_pdb_chain('1cbs')
- get_secondary_structure(query)[source]¶
Provides residue ranges of regular secondary structure
(alpha helices and beta strands) found in protein chains of the entry. For strands, sheet id can be used to identify a beta sheet.
- Parameters
query – a 4-character PDB id code
p.get_secondary_structure('1cbs')
- get_summary(query)[source]¶
Returns summary of a PDB entry
This can be title of the entry, list of depositors, date of deposition, date of release, date of latest revision, experimental method, list of related entries in case split entries, etc.
- Parameters
query – a 4-character PDB id code
p.get_summary('1cbs') p.get_summary('1cbs,2kv8') p.get_summary(['1cbs', '2kv8'])
8.25. PRIDE module¶
Interface to PRIDE web service
What is PRIDE ?
The PRIDE PRoteomics IDEntifications database is a centralized, standards compliant, public data repository for proteomics data, including protein and peptide identifications, post-translational modifications and supporting spectral evidence.
—From PRIDE web site, Jan 2015
- class PRIDE(verbose=False, cache=False)[source]¶
Interface to the PRIDE service
Constructor
- Parameters
verbose – set to False to prevent informative messages
- get_assay_count(identifier)[source]¶
Count assays for a project accession number
- Parameters
identifier (str) – a project accession number
- Returns
integer
>>> p = PRIDE() >>> assays = p.get_assay_count('PRD000001') 5
- get_assay_list(identifier)[source]¶
Return list of assays for a project accession nuber
- Parameters
identifier (str) – project accession number. See
get_project_list()- Returns
list of dictionaries. Each dictionary represents an assay.
>>> p = PRIDE() >>> assays = p.get_assay_list('PRD000001') >>> len(assays) # could be found with get_assay_count_project_accession 5 >>> assays[1]['assayAccession'] 1643
- get_assays(identifier)[source]¶
Retrieve assay information by assay accession
- Parameters
identifier (int) – assay accession number
>>> p = PRIDE() >>> res = p.get_assays(1643) >>> res['proteinCount'] 276
- get_file_count(identifier)[source]¶
return count of files in a project
- Parameters
identifier (str) – a project accession number
- Returns
int
>>> p.get_file_count('PRD000001') 5
- get_file_count_assay(identifier)[source]¶
list files for an assay
- Parameters
identifier (int) – assay accession number
- Returns
int
p.get_file_assay(1643)
- get_file_list(identifier)[source]¶
return list of files for a project
- Parameters
identifier (str) – a project accession number
>>> files = p.get_file_count('PRD000001') >>> len(files) 5
- get_file_list_assay(identifier)[source]¶
list files for an assay
- Parameters
identifier (int) – assay accession number
- Returns
list of dictionary, Each dictionary represents a file data structure
res = p.get_file_assay(1643)
- get_peptide_count(identifier, sequence=None)[source]¶
Count peptide identifications by project accession
- Parameters
identifier (str) – a project accession number
- Returns
int
>>> p.get_peptide_count('PRD000001', sequence='PLIPIVVEQTGR') 4 >>> p.get_peptide_count('PRD000001') 6758
- get_peptide_count_assay(identifier, sequence=None)[source]¶
Count peptide identifications by assay accession
- Parameters
identifier (str) – an assay accession number
- Returns
int
>>> p.get_peptide_count_assay(1643, sequence='AAATQKKVER') 5 >>> p.get_peptide_count_assay(1643) 1696
- get_peptide_list(identifier, sequence=None, show=10, page=0)[source]¶
Retrieve peptide identifications by project accession (and sequence)
- Parameters
>>> peptides = p.get_peptide_list('PRD000001', sequence='PLIPIVVEQTGR') >>> len(peptides) 4 >>> peptides = p.get_peptide_list('PRD000001') >>> len(peptides) 10 >>> peptides = p.get_peptide_list('PRD000001', show=100)
Note
the function merge two functions from the PRIDE API (get_peptide_list and get_peptide_list_sequence)
- get_peptide_list_assay(identifier, sequence=None, show=10, page=0)[source]¶
Retrieve peptide identifications by assay accession (and sequence)
- Parameters
>>> peptides = p.get_peptide_list_assay(1643, sequence='AAATQKKVER') >>> len(peptides) 5 >>> peptides = p.get_peptide_list_assay(1643) >>> len(peptides) 10 >>> peptides = p.get_peptide_list_assay(1643, show=100)
Note
the function merge two functions from the PRIDE API (get_peptide_list and get_peptide_list_sequence)
- get_project(identifier)[source]¶
Retrieve project information by accession
- Parameters
identifier (str) – a valid PRIDE identifier e.g., PRD000001
- Returns
a dictionary with the project details. See http://www.ebi.ac.uk/pride/ws/archive/#!/project for details
>>> from bioservices import PRIDE >>> p = PRIDE() >>> res = p.get_project("PRD000001") >>> res['numPeptides'] 6758
- get_project_count(query='', speciesFilter=None, ptmsFilter=None, tissueFilter=None, diseaseFilter=None, titleFilter=None, instrumentFilter=None, experimentTypeFilter=None, quantificationfilter=None, projectTagFilter=None)[source]¶
Count projects for given criteria
Takes same query parameters as the /list operation; typically used to retrieve number of results before querying with /list
- Parameters
query (str) – search term to query for
speciesFilter (str) – filter by species (NCBI taxon ID or name)
ptmsFilter (str) – filter by PTM annotation query
tissueFilter (str) – filter by tissue annotation
diseaseFilter (str) – filter by disease annotation
titleFilter (str) – filter the title for keywords
instrumentFilter (str) – filter for instrument names or keywords
experimentTypeFilter (str) – filter by experiment type
quantificationFilter (str) – filter by quantification annotation
projectTagFilter (str) – filter by project tags
- Returns
number of projects (integer)
- get_project_list(query='', show=10, page=0, sort=None, order='desc', speciesFilter=None, ptmsFilter=None, tissueFilter=None, diseaseFilter=None, titleFilter=None, instrumentFilter=None, experimentTypeFilter=None, quantificationfilter=None, projectTagFilter=None)[source]¶
list projects or given criteria
- Parameters
query (str) – search term to query for
show (int) – how many results to return per page
page (int) – which page (starting from 0) of the result to return
sort (str) – the field to sort on
order (str) – the sorting order (asc or desc)
speciesFilter (str) – filter by species (NCBI taxon ID or name)
ptmsFilter (str) – filter by PTM annotation query
tissueFilter (str) – filter by tissue annotation
diseaseFilter (str) – filter by disease annotation
titleFilter (str) – filter the title for keywords
instrumentFilter (str) – filter for instrument names or keywords
experimentTypeFilter (str) – filter by experiment type
quantificationFilter (str) – filter by quantification annotation
projectTagFilter (str) – filter by project tags
>>> p = PRIDE() >>> projects = p.get_project_list(show=100)
- get_protein_count(identifier)[source]¶
Count protein identifications by project accession
- Parameters
identifier (str) – a project accession number
- Returns
int
- get_protein_count_assay(identifier)[source]¶
Count protein identifications by assay accession
- Parameters
identifier (str) – a project accession number
- Returns
int
8.26. PSICQUIC¶
Interface to the PSICQUIC web service
What is PSICQUIC ?
“PSICQUIC is an effort from the HUPO Proteomics Standard Initiative (HUPO-PSI) to standardise the access to molecular interaction databases programmatically. The PSICQUIC View web interface shows that PSICQUIC provides access to 25 active service “
—Dec 2012
8.26.1. About queries¶
source: PSICQUIC View web page
The idea behind PSICQUIC is to retrieve information related to protein interactions from various databases. Note that protein interactions does not necesseraly mean protein-protein interactions. In order to be effective, the query format has been standarised.
To do a search you can use the Molecular Interaction Query Language which is based on Lucene’s syntax. Here are some rules
Use OR or space ‘ ‘ to search for ANY of the terms in a field
Use AND if you want to search for those interactions where ALL of your terms are found
Use quotes (”) if you look for a specific phrase (group of terms that must be searched together) or terms containing special characters that may otherwise be interpreted by our query engine (eg. ‘:’ in a GO term)
Use parenthesis for complex queries (e.g. ‘(XXX OR YYY) AND ZZZ’)
- Wildcards (*,?) can be used between letters in a term or at the end of terms to do fuzzy queries,
but never at the beginning of a term.
- Optionally, you can prepend a symbol in front of your term.
(plus): include this term. Equivalent to AND. e.g. +P12345
(minus): do not include this term. Equivalent to NOT. e.g. -P12345
Nothing in front of the term. Equivalent to OR. e.g. P12345
Implicit fields are used when no field is specified (simple search). For instance, if you put ‘P12345’ in the simple query box, this will mean the same as identifier:P12345 OR pubid:P12345 OR pubauth:P12345 OR species:P12345 OR type:P12345 OR detmethod:P12345 OR interaction_id:P12345
8.26.2. About the MITAB output¶
The output returned by a query contains a list of entries. Each entry is formatted following the MITAB output.
Here below are listed the name of the field returned ordered as they would appear in one entry. The first item is always idA whatever version of MITAB is used. The version 25 of MITAB contains the first 15 fields in the table below. Newer version may incude more fields but always include the 15 from MITAB 25 in the same order. See the link from irefindex about mitab for more information.
Field Name |
Searches on |
Implicit* |
Example |
|---|---|---|---|
idA |
Identifier A |
No |
idA:P74565 |
idB |
Identifier B |
No |
idB:P74565 |
id |
Identifiers (A or B) |
No |
id:P74565 |
alias |
Aliases (A or B) |
No |
alias:(KHDRBS1 HCK) |
identifiers |
Identifiers and Aliases undistinctively |
Yes |
identifier:P74565 |
pubauth |
Publication 1st author(s) |
Yes |
pubauth:scott |
pubid |
Publication Identifier(s) OR |
Yes |
pubid:(10837477 12029088) |
taxidA |
Tax ID interactor A: the tax ID or the species name |
No |
taxidA:mouse |
taxidB |
Tax ID interactor B: the tax ID or species name |
No |
taxidB:9606 |
species |
Species. Tax ID A or Tax ID B |
Yes |
species:human |
type |
Interaction type(s) |
Yes |
type:”physical interaction” |
detmethod |
Interaction Detection method(s) |
Yes |
detmethod:”two hybrid*” |
interaction_id |
Interaction identifier(s) |
Yes |
interaction_id:EBI-761050 |
pbioroleA |
Biological role A |
Yes |
pbioroleA:ancillary |
pbioroleB |
Biological role B |
Yes |
pbioroleB:”MI:0684” |
pbiorole |
Biological roles (A or B) |
Yes |
pbiorole:enzyme |
ptypeA |
Interactor type A |
Yes |
ptypeA:protein |
ptypeB |
Interactor type B |
Yes |
ptypeB:”gene” |
ptype |
Interactor types (A or B) |
Yes |
pbiorole:”small molecule” |
pxrefA |
Interactor xref A (or Identifier A) |
Yes |
pxrefA:”GO:0003824” |
pxrefB |
Interactor xref B (or Identifier B) |
Yes pxrefB:”GO:0003824” |
|
pxref |
Interactor xrefs (A or B or Identifier A or Identifier B) |
Yes |
pxref:”catalytic activity” |
xref |
Interaction xrefs (or Interaction identifiers) |
Yes |
xref:”nuclear pore” |
annot |
Interaction annotations and tags |
Yes |
annot:”internally curated” |
udate |
Update date |
Yes |
udate:[20100101 TO 20120101] |
negative |
Negative interaction boolean |
Yes |
negative:true |
complex |
Complex expansion |
Yes |
complex:”spoke expanded” |
ftypeA |
Feature type of participant A |
Yes |
ftypeA:”sufficient to bind” |
ftypeB |
Feature type of participant B |
Yes |
ftypeB:mutation |
ftype |
Feature type of participant A or B |
Yes |
ftype:”binding site” |
pmethodA |
Participant identification method A |
Yes |
pmethodA:”western blot” |
pmethodB |
Participant identification method B |
Yes |
pmethodB:”sequence tag identification” |
pmethod |
|
Yes |
pmethod:immunostaining |
stc |
Stoichiometry (A or B). Only true or false, just to be able to filter interaction having stoichiometry available |
Yes |
stc:true |
param |
Interaction parameters. Only true or false, just to be able to filter interaction having parameters available |
Yes |
param:true |
- class PSICQUIC(verbose=True)[source]¶
Interface to the PSICQUIC service
There are 2 interfaces to the PSICQUIC service (REST and WSDL) but we used the REST only.
This service provides a common interface to more than 25 other services related to protein. So, we won’t detail all the possiblity of this service. Here is an example that consists of looking for interactors of the protein ZAP70 within the IntAct database:
>>> from bioservices import * >>> s = PSICQUIC() >>> res = s.query("intact", "zap70") >>> len(res) # there are 11 interactions found 11 >>> for x in res[1]: ... print(x) uniprotkb:O95169 uniprotkb:P43403 intact:EBI-716238 intact:EBI-1211276 psi-mi:ndub8_human(display_long)|uniprotkb:NADH-ubiquinone oxidoreductase ASHI . .
Here we have a list of entries. There are 15 of them (depending on the output parameter). The meaning of the entries is described on PSICQUIC website: https://code.google.com/p/psicquic/wiki/MITAB25Format . In short:
Unique identifier for interactor A
Unique identifier for interactor B.
Alternative identifier for interactor A, for example the official gene
Alternative identifier for interactor B.
Aliases for A, separated by “|
Aliases for B.
Interaction detection methods, taken from the corresponding PSI-MI
First author surname(s) of the publication(s)
Identifier of the publication
NCBI Taxonomy identifier for interactor A.
NCBI Taxonomy identifier for interactor B.
Interaction types,
Source databases and identifiers,
Interaction identifier(s) i
Confidence score. Denoted as scoreType:value.
Another example with reactome database:
res = s.query("reactome", "Q9Y266")
Warning
PSICQUIC gives access to 25 other services. We cannot create a dedicated parsing for all of them. So, the ::query method returns the raw data. Addition class may provide dedicated parsing in the future.
See also
Constructor
- Parameters
verbose (bool) – print informative messages
>>> from bioservices import PSICQUIC >>> s = PSICQUIC()
- property activeDBs¶
returns the active DBs only
- property formats¶
Returns the possible output formats
- getInteractionCounter(query)[source]¶
Returns a dictionary with database as key and results as values
- Parameters
query (str) – a valid query
- Returns
a dictionary which key as database and value as number of entries
Consider only the active database.
- knownName(data)[source]¶
Scan all entries (MITAB) and returns simplified version
Each item in the input list of mitab entry The output is made of 2 lists corresponding to interactor A and B found in the mitab entries.
elements in the input list takes the following forms:
DB1:ID1|DB2:ID2 DB3:ID3
The | sign separates equivalent IDs from different databases.
We want to keep only one. The first known databae is kept. If in the list of DB:ID pairs no known database is found, then we keep the first one whatsover.
known databases are those available in the uniprot mapping tools.
chembl and chebi IDs are kept unchanged.
- postCleaning(data, keep_only='HUMAN', remove_db=['chebi', 'chembl'], keep_self_loop=False, verbose=True)[source]¶
Remove entries with a None and keep only those with the keep pattern
- postCleaningAll(data, keep_only='HUMAN', flatten=True, verbose=True)[source]¶
even more cleaing by ignoring score, db and interaction len(set([(x[0],x[1]) for x in retnew]))
- print_status()[source]¶
Prints the services that are available
- Returns
Nothing
The output is tabulated. The columns are:
names
active
count
version
rest URL
soap URL
rest example
restricted
See also
If you want the data into lists, see all attributes starting with registry such as
registry_names()
- query(service, query, output='tab25', version='current', firstResult=None, maxResults=None)[source]¶
Send a query to a specific database
- Parameters
service (str) – a registered service. See
registry_names.query (str) – a valid query. Can be * or a protein name.
output (str) – a valid format. See s._formats
s.query("intact", "brca2", "tab27") s.query("intact", "zap70", "xml25") s.query("matrixdb", "*", "xml25")
This is the programmatic approach to this website:
http://www.ebi.ac.uk/Tools/webservices/psicquic/view/main.xhtml
Another example consist in accessing the string database for fetching protein-protein interaction data of a particular model organism. Here we restrict the query to 100 results:
s.query("string", "species:10090", firstResult=0, maxResults=100, output="tab25")
# spaces are automatically converted
s.query(“biogrid”, “ZAP70 AND species:9606”)
Warning
AND must be in big caps. Some database are ore permissive than other (e.g., intact accepts “and”). species must be a valid ID number. Again, some DB are more permissive and may accept the name (e.g., human)
To obtain the number of interactions in intact for the human specy:
>>> len(p.query("intact", "species:9606"))
- queryAll(query, databases=None, output='tab25', version='current', firstResult=None, maxResults=None)[source]¶
Same as query but runs on all active database
- Parameters
databases (list) – database to query. Queries all active DB if not provided
- Returns
dictionary where keys correspond to databases and values to the output of the query.
res = s.queryAll("ZAP70 AND species:9606")
- property registry¶
returns the registry of psicquic
- property registry_actives¶
returns active state of each service
- property registry_counts¶
returns number of entries in each service
- property registry_names¶
returns all services available (names)
- property registry_restexamples¶
retuns REST example for each service
- property registry_restricted¶
returns restricted status of services
- property registry_resturls¶
returns URL of REST services
- property registry_soapurls¶
returns URL of WSDL service
- property registry_versions¶
returns version of each service
8.27. Rhea¶
Interface to the Rhea web services
What is Rhea ?
- URL
- Citations
Rhea is a reaction database, where all reaction participants (reactants and products) are linked to the ChEBI database (Chemical Entities of Biological Interest) which provides detailed information about structure, formula and charge. Rhea provides built-in validations that ensure both elemental and charge balance of the reactions… While the main focus of Rhea is enzyme-catalysed reactions, other biochemical reactions are also are included.
The database is extensively cross-referenced. Reactions are currently linked to the EC list, KEGG and MetaCyc, and the reactions will be used in the IntEnz database and in all relevant UniProtKB entries. Furthermore, the reactions will also be used in the UniPathway database to generate pathways and metabolic networks.
—from Rhea Home page, Dec 2012 (http://www.ebi.ac.uk/rhea/about.xhtml)
- class Rhea(verbose=True, cache=False)[source]¶
Interface to the Rhea service
You can search by compound name, ChEBI ID, reaction ID, cross reference (e.g., EC number) or citation (author name, title, abstract text, publication ID). You can use double quotes - to match an exact phrase - and the following wildcards:
? (question mark = one character),
* (asterisk = several characters).
Searching for caffe* will find reactions with participants such as caffeine, trans-caffeic acid or caffeoyl-CoA:
from bioservices import Rhea r = Rhea() response = r.search("caffe*")
Searching for a?e?o* will find reactions with participants such as acetoin, acetone or adenosine.:
from bioservices import Rhea r = Rhea() response = r.search("a?e?o*")
The
search()entry()methods require a list of valid columns. By default all columns are used but you can restrict to only a few. Here is the description of the columns:rhea-id : reaction identifier (with prefix RHEA) equation : textual description of the reaction equation chebi : comma-separated list of ChEBI names used as reaction participants chebi-id : comma-separated list of ChEBI identifiers used as reaction participants ec : comma-separated list of EC numbers (with prefix EC) uniprot : number of proteins (UniProtKB entries) annotated with the Rhea reaction pubmed : comma-separated list of PubMed identifiers (without prefix)
and 5 cross-references:
reaction-xref(EcoCyc) reaction-xref(MetaCyc) reaction-xref(KEGG) reaction-xref(Reactome) reaction-xref(M-CSA)
Rhea constructor
- Parameters
verbose (bool) – True by default
>>> from bioservices import Rhea >>> r = Rhea()
- get_metabolites(rxn_id)[source]¶
Given a Rhea (http://www.rhea-db.org/) reaction id, returns its participant metabolites as a dict: {metabolite: stoichiometry},
e.g. ‘2 H + 1 O2 = 1 H2O’ would be represented ad {‘H’: -2, ‘O2’: -1, ‘H2O’: 1}.
- Parameters
rxn_id – Rhea reaction id
- Returns
dict of participant metabolites.
- query(query, columns=None, frmt='tsv', limit=None)[source]¶
Retrieve a concrete reaction for the given id in a given format
- Parameters
- Returns
dataframe
Retrieve Rhea reaction identifiers and equation text:
r.query("", columns="rhea-id,equation", limit=10)
Retrieve Rhea reactions with enzymes curated in UniProtKB (only first 10 entries):
r.query("uniprot:*", columns="rhea-id,equation", limit=10)
To retrieve a specific entry:
df = r.get_entry("rhea:10661")
Changed in version 1.8.0: (entry() method renamed in query() and no more format required. Must be given in the entry name e.g. query(“10281.rxn”) instead of entry(10281, format=”rxn”) the option frmt is now related to the result format
- search(query, columns=None, limit=None, frmt='tsv')[source]¶
Search for Rhea (mimics https://www.rhea-db.org/)
- Parameters
- Returns
A pandas DataFrame.
>>> r = Rhea() >>> df = r.search("caffeine") >>> df = r.search("caffeine", columns='rhea-id,equation')
8.28. Reactome¶
Interface to the Reactome webs services
What is Reactome?
- URL
- Citation
- REST
http://reactomews.oicr.on.ca:8080/ReactomeRESTfulAPI/RESTfulWS
“REACTOME is an open-source, open access, manually curated and peer-reviewed pathway database. Pathway annotations are authored by expert biologists, in collaboration with Reactome editorial staff and cross-referenced to many bioinformatics databases. These include NCBI Entrez Gene, Ensembl and UniProt databases, the UCSC and HapMap Genome Browsers, the KEGG Compound and ChEBI small molecule databases, PubMed, and Gene Ontology. … “
—from Reactome web site
- class Reactome(verbose=True, cache=False)[source]¶
Todo
interactors, orthology, particiapnts, person, query, refernces, schema
- get_complex_subunits(identifier, excludeStructuresSpecifies=False)[source]¶
A list with the entities contained in a given complex
Retrieves the list of subunits that constitute any given complex. In case the complex comprises other complexes, this method recursively traverses the content returning each contained PhysicalEntity. Contained complexes and entity sets can be excluded setting the ‘excludeStructures’ optional parameter to ‘true’
- Parameters
identifier – The complex for which subunits are requested
excludeStructures – Specifies whether contained complexes and entity sets are excluded in the response
r.get_complex_subunits("R-HSA-5674003")
- get_complexes(resources, identifier)[source]¶
A list of complexes containing the pair (identifier, resource)
Retrieves the list of complexes that contain a given (identifier, resource). The method deconstructs the complexes into all its participants to do so.
- Parameters
resource – The resource of the identifier for complexes are requested (e.g. UniProt)
identifier – The identifier for which complexes are requested
r.get_complexes(resources, identifier) r.get_complexes("UniProt", "P43403")
- get_discover(identifier)[source]¶
The schema.org for an Event in Reactome knowledgebase
For each event (reaction or pathway) this method generates a json file representing the dataset object as defined by schema.org (http). This is mainly used by search engines in order to index the data
r.data_discover("R-HSA-446203")
- get_diseases_doid()[source]¶
retrieves the list of disease DOIDs annotated in Reactome
return: dictionary with DOID contained in the values()
- get_entity_componentOf(identifier)[source]¶
A list of larger structures containing the entity
Retrieves the list of structures (Complexes and Sets) that include the given entity as their component. It should be mentioned that the list includes only simplified entries (type, names, ids) and not full information about each item.
r.get_entity_componentOf("R-HSA-199420")
- get_entity_otherForms(identifier)[source]¶
All other forms of PhysicalEntity
Retrieves a list containing all other forms of the given PhysicalEntity. These other forms are PhysicalEntities that share the same ReferenceEntity identifier, e.g. PTEN H93R[R-HSA-2318524] and PTEN C124R[R-HSA-2317439] are two forms of PTEN.
r.get_entity_otherForms("R-HSA-199420")
- get_event_ancestors(identifier)[source]¶
The ancestors of a given event
The Reactome definition of events includes pathways and reactions. Although events are organised in a hierarchical structure, a single event can be in more than one location, i.e. a reaction can take part in different pathways while, in the same way, a sub-pathway can take part in many pathways. Therefore, this method retrieves a list of all possible paths from the requested event to the top level pathway(s).
- Parameters
identifier – The event for which the ancestors are requested
r.get_event_ancestors("R-HSA-5673001")
- get_eventsHierarchy(species)[source]¶
The full event hierarchy for a given species
Events (pathways and reactions) in Reactome are organised in a hierarchical structure for every species. By following all ‘hasEvent’ relationships, this method retrieves the full event hierarchy for any given species. The result is a list of tree structures, one for each TopLevelPathway. Every event in these trees is represented by a PathwayBrowserNode. The latter contains the stable identifier, the name, the species, the url, the type, and the diagram of the particular event.
- Parameters
species – Allowed species filter: SpeciesName (eg: Homo sapiens) SpeciesTaxId (eg: 9606)
r.get_eventsHierarchy(9606)
- get_exporter_diagram(identifier, ext='png', quality=5, diagramProfile='Modern', analysisProfile='Standard', filename=None)[source]¶
Export a given pathway diagram to raster file
This method accepts identifiers for Event class instances. When a diagrammed pathway is provided, the diagram is exported to the specified format. When a subpathway is provided, the diagram for the parent is exported and the events that are part of the subpathways are selected. When a reaction is provided, the diagram containing the reaction is exported and the reaction is selected.
- Parameters
identifier – Event identifier (it can be a pathway with diagram, a subpathway or a reaction)
ext – File extension (defines the image format) in png, jpeg, jpg, svg, gif
quality – Result image quality between [1 - 10]. It defines the quality of the final image (Default 5)
flg – not implemented
sel – not implemented
diagramProfile – Diagram Color Profile
token – not implemented
analysisProfile – Analysis Color Profile
expColumn – not implemented
filename – if given, save the results in the provided filename
- return: raw data if filename parameter is not set. Otherwise, the data
is saved in the filename and the function returns None
- get_exporter_sbml(identifier)[source]¶
Export given Pathway to SBML
- Parameters
identifier – DbId or StId of the requested database object
r.exporter_sbml("R-HSA-68616")
- get_interactors_psicquic_molecule_details()[source]¶
Retrieve clustered interaction, sorted by score, of a given accession by resource.
- get_interactors_psicquic_molecule_summary()[source]¶
Retrieve a summary of a given accession by resource
- get_interactors_static_molecule_details()[source]¶
Retrieve a detailed interaction information of a given accession
- get_interactors_static_molecule_pathways()[source]¶
Retrieve a list of lower level pathways where the interacting molecules can be found
- get_pathway_containedEvents(identifier)[source]¶
All the events contained in the given event
Events are the building blocks used in Reactome to represent all biological processes, and they include pathways and reactions. Typically, an event can contain other events. For example, a pathway can contain smaller pathways and reactions. This method recursively retrieves all the events contained in any given event.
res = r.get_pathway_containedEvents("R-HSA-5673001")
- get_pathway_containedEvents_by_attribute(identifier, attribute)[source]¶
A single property for each event contained in the given event
Events are the building blocks used in Reactome to represent all biological processes, and they include pathways and reactions. Typically, an event can contain other events. For example, a pathway can contain smaller pathways (subpathways) and reactions. This method recursively retrieves a single attribute for each of the events contained in the given event.
- Parameters
identifier – The event for which the contained events are requested
attribute – Attrubute to be filtered
r.get_pathway_containedEvents_by_attribute("R-HSA-5673001", "stId")
- get_pathways_low_diagram_entity(identifier)[source]¶
A list of lower level pathways with diagram containing a given entity or event
This method traverses the event hierarchy and retrieves the list of all lower level pathways that have a diagram and contain the given PhysicalEntity or Event.
- Parameters
identifier – The entity that has to be present in the pathways
species – The species for which the pathways are requested. Taxonomy identifier (eg: 9606) or species name (eg: ‘Homo sapiens’)
r.get_pathways_low_diagram_entity("R-HSA-199420")
- get_pathways_low_diagram_entity_allForms(identifier)[source]¶
r.get_pathways_low_diagram_entity_allForms("R-HSA-199420")
- get_pathways_low_entity(identifier)[source]¶
A list of lower level pathways containing a given entity or event
This method traverses the event hierarchy and retrieves the list of all lower level pathways that contain the given PhysicalEntity or Event.
r.get_pathways_low_entity("R-HSA-199420")
- get_pathways_low_entity_allForms(identifier)[source]¶
A list of lower level pathways containing any form of a given entity
This method traverses the event hierarchy and retrieves the list of all lower level pathways that contain the given PhysicalEntity in any of its variant forms. These variant forms include for example different post-translationally modified versions of a single protein, or the same chemical in different compartments.
r.get_pathways_low_entity_allForms("R-HSA-199420")
- get_references(identifier)[source]¶
All referenceEntities for a given identifier
Retrieves a list containing all the reference entities for a given identifier.
r.get_references(15377)
- property name¶
- search_facet()[source]¶
A list of facets corresponding to the whole Reactome search data
This method retrieves faceting information on the whole Reactome search data.
- search_facet_query(query)[source]¶
A list of facets corresponding to a specific query
This method retrieves faceting information on a specific query
- search_query(query)[source]¶
Queries Solr against the Reactome knowledgebase
This method performs a Solr query on the Reactome knowledgebase. Results can be provided in a paginated format.
- search_spellcheck(query)[source]¶
Spell-check suggestions for a given query
This method retrieves a list of spell-check suggestions for a given search term.
- search_suggest(query)[source]¶
Autosuggestions for a given query
This method retrieves a list of suggestions for a given search term.
>>> r.http_get("search/suggest?query=apopt") ['apoptosis', 'apoptosome', 'apoptosome-mediated', 'apoptotic']
- property version¶
- class ReactomeAnalysis(verbose=True, cache=False)[source]¶
Constructor
- Parameters
name (str) – a name for this service
url (str) – its URL
verbose (bool) – prints informative messages if True (default is True)
requests_per_sec – maximum number of requests per seconds are restricted to 3. You can change that value. If you reach the limit, an error is raise. The reason for this limitation is that some services (e.g.., NCBI) may black list you IP. If you need or can do more (e.g., ChEMBL does not seem to have restrictions), change the value. You can also have several instance but again, if you send too many requests at the same, your future requests may be retricted. Currently implemented for REST only
All instances have an attribute called
loggingthat is an instanceof theloggingmodule. It can be used to print information, warning, error messages:self.logging.info("informative message") self.logging.warning("warning message") self.logging.error("error message")
The attribute
debugLevelcan be used to set the behaviour of the logging messages. If the argument verbose is True, the debugLebel is set to INFO. If verbose if False, the debugLevel is set to WARNING. However, you can use thedebugLevelattribute to change it to one of DEBUG, INFO, WARNING, ERROR, CRITICAL. debugLevel=WARNING means that only WARNING, ERROR and CRITICAL messages are shown.
- class ReactomeOld(verbose=True, cache=False)[source]¶
Reactome interface
some data can be download on the main website
Constructor
- Parameters
name (str) – a name for this service
url (str) – its URL
verbose (bool) – prints informative messages if True (default is True)
requests_per_sec – maximum number of requests per seconds are restricted to 3. You can change that value. If you reach the limit, an error is raise. The reason for this limitation is that some services (e.g.., NCBI) may black list you IP. If you need or can do more (e.g., ChEMBL does not seem to have restrictions), change the value. You can also have several instance but again, if you send too many requests at the same, your future requests may be retricted. Currently implemented for REST only
All instances have an attribute called
loggingthat is an instanceof theloggingmodule. It can be used to print information, warning, error messages:self.logging.info("informative message") self.logging.warning("warning message") self.logging.error("error message")
The attribute
debugLevelcan be used to set the behaviour of the logging messages. If the argument verbose is True, the debugLebel is set to INFO. If verbose if False, the debugLevel is set to WARNING. However, you can use thedebugLevelattribute to change it to one of DEBUG, INFO, WARNING, ERROR, CRITICAL. debugLevel=WARNING means that only WARNING, ERROR and CRITICAL messages are shown.- SBML_exporter(identifier)[source]¶
Get the SBML XML text of a pathway identifier
- Parameters
identifier (int) – Pathway database identifier
- Returns
SBML object in XML format as a string
>>> from bioservices import Reactome >>> s = Reactome() >>> xml = s.SBML_exporter(109581)
- biopax_exporter(identifier, level=2)[source]¶
Get BioPAX file
The passed identifier has to be a valid event identifier. If there is no matching ID in the database, it will return an empty string.
- Parameters
- Returns
BioPAX RDF document
>>> # for Apoptosis: >>> s = Reactome() >>> res = s.biopax_exporter(109581)
- bioservices_get_reactants_from_reaction_identifier(reaction)[source]¶
Fetch information from the reaction HTML page
Note
draft version
- front_page_items(species)[source]¶
Get list of front page items listed in the Reactome Pathway Browser
- Parameters
species (str) – Full species name that should be encoded for URL (e.g. homo+sapiens for human, or mus+musculus for mouse) + can be replaced by spaces.
- Returns
list of fully encoded Pathway objects in JSON
>>> s = Reactome() >>> res = s.front_page_items("homo sapiens") >>> print(res[0]['name']) ['Apoptosis']
See also
- get_list_pathways()[source]¶
Return list of pathways from reactome website
- Returns
list of lists. Each sub-lis contains 3 items: reactome pathway identifier, description and species
- highlight_pathway_diagram(identifier, genes, frmt='PNG')[source]¶
Highlight a diagram for a specified pathway based on its identifier
- Parameters
- Returns
This method should be used after method queryHitPathways.
res = s.http_post("highlightPathwayDiagram/68875/PNG", frmt="txt", data="CDC2") with open("test.png", 'wb') as f: import binascii f.write(binascii.a2b_base64(res))
- list_by_query(classname, **kargs)[source]¶
Get list of objecs from Reactome database
- Parameters
name (str class) –
kargs – further attribute values encoded in key-value pair
- Returns
list of dictionaries. Each dictionary contains information about a given pathway
To query a list of pathways with names as “Apoptosis”:
>>> s = Reactome() >>> res = list_by_query("Pathway", name="apoptosis") >>> identifiers = [x['dbId'] for x in res]
- pathway_complexes(identifier)[source]¶
Get complexes belonging to a pathway
- Parameters
identifier (int) – Pathway database identifier
- Returns
list of all PhysicalEntity objects that participate in the Pathway.(in JSON)
>>> s = Reactome() >>> s.pathway_complexes(109581)
- pathway_diagram(identifier, frmt='PNG')[source]¶
Retrieve pathway diagram
- Parameters
- Returns
Base64 encoded pathway diagram for PNG or PDF. XML text for the XML file type.
>>> s = Reactome() >>> s.pathway_diagram('109581', 'PNG',view=True) >>> s.pathway_diagram('109581', 'PNG', save=True)
Todo
if PNG or PDF, the output is base64 but there is no facility to easily save the results in a file for now
- pathway_hierarchy(species)[source]¶
Get the pathway hierarchy for a species as displayed in Reactome pathway browser.
- Parameters
species (str) – species name that should be with + or spaces (e.g. ‘homo+sapiens’ for human, or ‘mus musculus’ for mouse)
- Returns
XML text containing pathways and reactions
s.pathway_hierarchy("homo sapiens")
- pathway_participants(identifier)[source]¶
Get list of pathway participants for a pathway using
- Parameters
identifier (int) – Pathway database identifier
- Returns
list of fully encoded PhysicalEntity objects in the pathway (in JSON)
>>> s = Reactome() >>> s.pathway_participants(109581)
- query_by_id(classname, identifier)[source]¶
Get Reactome Database for a specific object.
- Parameters
It returns a full object, including full class information about all the attributes of the returned object. For example, if the object has one PublicationSource attribute, it will return a full PublicationSource object within the returned object.
>>> s.query_by_id("Pathway", "109581")
- query_by_ids(classname, identifiers)[source]¶
-
>>> s.quey_by_ids("Pathway", "CDC2")
Warning
not sure the wrapping is correct
- query_hit_pathways(query)[source]¶
Get pathways that contain one or more genes passed in the query list.
In the Reactome data model, pathways are organized in a hierarchical structure. The returned pathways in this method are pathways having detailed manually drawn pathway diagrams. Currently only human pathways will be returned from this method.
s.query_hit_pathways('CDC2') s.query_hit_pathways(['CDC2'])
8.29. Readseq¶
This module provides a class Seqret to access to Seqret WS.
What is Seqret ?
- URL
- Service
- Citations
EMBOSS seqret reads and converts biosequences between a selection of common biological sequence formats, including EMBL, GenBank and fasta sequence formats.
Seqret homepage – Sep 2017
- class Seqret(verbose=True)[source]¶
Interface to the Seqret service
>>> from bioservices import * >>> s = Seqret()
The ReadSeq service was replaced by #the Seqret services (2015).
Changed in version 0.15.
Constructor
- Parameters
verbose (bool) –
- get_parameter_details(parameterId)[source]¶
Get details of a specific parameter.
- Parameters
parameter (str) – identifier/name of the parameter to fetch details of.
- Returns
a data structure describing the parameter and its values.
rs = ReadSeq() print(rs.get_parameter_details("stype"))
- get_parameters()[source]¶
Get a list of the parameter names.
- Returns
a list of strings giving the names of the parameters.
- get_result(jobid, result_type='out')[source]¶
Get the result of a job of the specified type.
- Parameters
jobid (str) – job identifier.
parameters – optional list of wsRawOutputParameter used to provide additional parameters for derived result types.
- get_result_types(jobid)[source]¶
Get the available result types for a finished job.
- Parameters
jobid (str) – job identifier.
- Returns
a list of wsResultType data structures describing the available result types.
- get_status(jobid=None)[source]¶
Get the status of a submitted job.
- Parameters
jobid (str) – job identifier.
- Returns
string containing the status.
The values for the status are:
RUNNING: the job is currently being processed.
FINISHED: job has finished, and the results can then be retrieved.
ERROR: an error occurred attempting to get the job status.
FAILURE: the job failed.
NOT_FOUND: the job cannot be found.
- property parameters¶
Get list of parameter names
- run(email, title, **kargs)[source]¶
Submit a job to the service.
- Parameters
email (str) – user e-mail address.
title (str) – job title.
params – parameters for the tool as returned by
get_parameter_details().
- Returns
string containing the job identifier (jobId).
Deprecated (olf readseq service):
Format Name Value Auto-detected 0 EMBL 4 GenBank 2 Fasta(Pearson) 8 Clustal/ALN 22 ACEDB 25 BLAST 20 DNAStrider 6 FlatFeat/FFF 23 GCG 5 GFF 24 IG/Stanford 1 MSF 15 NBRF 3 PAUP/NEXUS 17 Phylip(Phylip4) 12 Phylip3.2 11 PIR/CODATA 14 Plain/Raw 13 SCF 21 XML 19
As output, you also have
Pretty 18
s = readseq.Seqret() jobid = s.run("cokelaer@test.co.uk", "test", sequence=fasta, inputformat=8, outputformat=2) genbank = s.get_result(s._jobid)
8.30. UniChem¶
This module provides a class UniChem
What is UniChem
“UniChem is a ‘Unified Chemical Identifier’ system, designed to assist in the rapid cross-referencing of chemical structures, and their identifiers, between databases (read more). “
—From UniChem web page June 2013
- class UniChem(verbose=False, cache=False)[source]¶
Interface to the UniChem service
>>> from bioservices import UniChem >>> u = UniChem()
There are lots of sources such as Chembl, Chebi, etc. You will probably need the identifiers of those sources. You can get all information about a source using these methods:
# Get information about a source u.get_source_info_by_name('chembl') u.get_source_info_by_id(10) u.get_id_from_name('chembl') u.get_all_src_ids()
but for developers, everything is contained in the
source_idsdictionary.The first important method provided by Unichem API is the
get_compounds(). For example, you can request all compounds related to the CHEMBL12 identifier from ChEMBL using:res = u.get_compounds('CHEMBL12', 'chembl') compounds = res['compounds'][0]
Note that the second argument is ‘chembl’ and lower/upper cases is important. All names are stored in
source_idstogether with their identifiers.You can use also
get_id_from_name()and get_name_from_id` if needed.Legacy methods are available:
get_compound_ids_from_src_id –> use get_compounds() get_src_compound_ids_from_inchikey –> replaced by get_compounds() get_all_src_ids() –> uses new API get_src_compound_ids_all_from_inchikey –> get_source_by_inchikey() get_verbose_src_compound_ids_from_inchikey –> get_sources_by_inchikey_verbose() get_structure –> uses new API get_compounds() and bioservices code get_structure_all –> dropped get_src_compound_id_url –> dropped. One can use the get_compounds() get_src_compound_ids_all_from_obsolete –> removed
get_src_compound_ids_from_src_compound_id –> removed; was obsolet get_src_compound_ids_all_from_src_compound_id –> remoed was already obsolet get_all_compound_ids_from_all_src_id –> removed. no more API get_mapping –> removed. no more API get_auxiliary_mappings –> removed. no more API
Most old functions can be replaced by a syntax such as:
res = u.get_compound('CHEMBL12', 'chembl') res['compounds'][0]
Constructor UniChem
- Parameters
verbose – set to False to prevent informative messages
- get_all_src_ids()[source]¶
Obtain all src_ids of sources available in UniChem
- Returns
list of ‘src_id’s.
uni.get_all_src_ids()
- get_compounds(compound, source_type)[source]¶
Get matched compounds information
- Parameters
- Returns
a list of matched compounds and their assigned sources
A legacy function allows you to retrieve a compound from its inchikey:
u.get_sources_by_inchikey('GZUITABIAKMVPG-UHFFFAOYSA-N')
However, this new function is faster presumably and allows you to do the same:
res = u.get_compounds('GZUITABIAKMVPG-UHFFFAOYSA-N', 'inchikey') res['compounds']
You can get the first element, from which inchi, sources, standardInchikey, uci can be extracted. The sources key contains all compound identifiers for each source:
res['compounds'][0]['uci'] res['compounds'][0]['sources']
Looks like there is always a single element in res[‘compounds’] but since it is a list, you must access to first element (unique) using [0] syntax.
- get_connectivity(compound, source_type)[source]¶
Fetch multiple source data sets for a given compound with common connectivity to a given id on the database source, InChI, InChIkey or UCI
- Parameters
compound (str) – InChI, InChIKey, Name, UCI or Compound Source ID (e.g. chembl)
source_type – uci, inchi, inchikey, sourceID
The returned dictionary contains 5 keys:
response: service response (‘Success’ if everything is right)
searchedCompound: the summary in terms of inchi, standardInchikey and uci
- sources: a dictionary with e.g. compoundID and name of the source.
A ‘comparison’ dictionary is also provided.
totalCompounds: number of searchedCompound entries
totalSources: number of sources entries
- get_id_from_name(name)[source]¶
Return the ID a a source given its name.
- Parameters
name (str) – a valid database name (e.g., chembl)
u.get_id_from_name("chembl")
- get_images(uci, filename=None)[source]¶
Return / create compound image
- Parameters
uci – the UCI of the compound
filename – optional file name to save the SVG+XML output
- Returns
the SVG+XML string
- get_inchi_from_inchikey(inchikey)[source]¶
Get a list of inchis given a valid inchikey.
- Parameters
inchikey – InChI Key to search. Unlike the rest API, you can also provide a list.
- Returns
a list of inchis matching the InChI Key provided. If input is a list, a dictionary is returned where keys are the inchikey input lists.
from bioservices import UniChem u = UniChem() res = u.get_inchi("AAOVKJBEBIDNHE-UHFFFAOYSA-N")
Note
this is a legacy function. introduced in v1.9 after unichem API update
- get_source_info_by_name(src_name)[source]¶
Description: Obtain all information on a source by querying with a source id
- Parameters
src_name (int) – valid identifiers can be found in
source_idse.g. chebi, chembl)- Returns
dictionary (or list of dictionaries) with following keys:
UCICount: number of entries
baseIdUrl: URL of the source
created: date of creation
description: a description of the content of the source
lastUpdated: last date of the update
name: the unique name for the source in UniChem, always lower case
nameLabel: A name for the source suitable for use as a ‘label’ for the source
nameLong: the full name of the source, as defined by the source
private: is it private or not ?
sourceID: the src_id for this source
srcDetails: details about the source
srcReleaseDate: release date of the source database
srcReleaseNumber: release number of the source
srcUrl: src_url (the main home page of the source)
updateComments: possible updates from this source
>>> res = get_source_by_name("chebi")
- get_sources()[source]¶
Returns all information about all sources used in Unichem
from bioservices import UniChem u = UniChem() res = u.get_sources_information() res['sources']
- get_sources_by_inchikey(inchikey)[source]¶
Get sources by inchikey
- Parameters
inchikey – InChI Key to search. Unlike the rest API, you can also provide a list.
- Returns
A list of sources for the provided InChIKey if input is a single string. a dictionary with keys as inchikey if input is a list.
Note
this is a legacy function. introduced in v1.9 after unichem API update
- get_sources_by_inchikey_verbose(inchikey)[source]¶
Get sources by inchikey
- Parameters
inchikey – InChI Key to search. Unlike the rest API, you can also provide a list.
- Returns
A list of sources for the provided InChIKey if input is a single string. a dictionary with keys as inchikey if input is a list.
Note
this is a legacy function. introduced in v1.9 after unichem API update
8.31. UniProt¶
Interface to some part of the UniProt web service
What is UniProt ?
- URL
- Citation
“The Universal Protein Resource (UniProt) is a comprehensive resource for protein sequence and annotation data. The UniProt databases are the UniProt Knowledgebase (UniProtKB), the UniProt Reference Clusters (UniRef), and the UniProt Archive (UniParc). The UniProt Metagenomic and Environmental Sequences (UniMES) database is a repository specifically developed for metagenomic and environmental data.”
—From Uniprot web site (help/about) , Dec 2012
- class UniProt(verbose=False, cache=False)[source]¶
Interface to the UniProt service
>>> from bioservices import UniProt >>> u = UniProt(verbose=False) >>> u.mapping("UniProtKB_AC-ID", "KEGG", query='P43403') defaultdict(<type 'list'>, {'P43403': ['hsa:7535']}) >>> res = u.search("P43403") # Returns sequence on the ZAP70_HUMAN accession Id >>> sequence = u.search("ZAP70_HUMAN", columns="sequence")
Changed in version 1.10: Uniprot update its service in June 2022. Changes were made in the bioservices API with small changes. User API is more or less the same. Main issues that may be faced are related to change of output column names. Please see the
_legacy_namesfor corresponding changes.Some notes about searches. The and and or are now upper cases. The organism and taxonomy fields are now organism_id and taxonomy_id
Constructor
- Parameters
verbose – set to False to prevent informative messages
cache – set to True to cache request
- get_df(entries, nChunk=100, organism=None, limit=10)[source]¶
Given a list of uniprot entries, this method returns a dataframe with all possible columns
- Parameters
entries – list of valid entry name. if list is too large (about >200), you need to split the list
chunk –
limit – limit number of entries per identifier to 10. You can set it to None to keep all entries but this will be very slow
- Returns
dataframe with indices being the uniprot id (e.g. DIG1_YEAST)
- get_fasta(uniprot_id)[source]¶
Returns FASTA string given a valid identifier
- Parameters
uniprot_id (str) – a valid identifier (e.g. P12345)
This is just an alias to
retrieve()when setting the format to ‘fasta’. Method kept for legacy.
- mapping(fr='UniProtKB_AC-ID', to='KEGG', query='P13368', polling_interval_seconds=3, max_waiting_time=100)[source]¶
This is an interface to the UniProt mapping service
- Parameters
fr – the source database identifier. See
valid_mapping.to – the targetted database identifier. See
valid_mapping.query – a string containing one or more IDs separated by a space It can also be a list of strings.
polling_interval_seconds – the number of seconds between each status check of the current job
max_waiting_time – the maximum number of seconds to wait for the final answer.
- Returns
a dictionary with two possible keys. The first one is ‘results’ with the from / to answers and the second one ‘failedIds’ with Ids that were not found
>>> u.mapping("UniProtKB_AC-ID", "KEGG", 'P43403') {'results': [{'from': 'P43403', 'to': 'hsa:7535'}]}
The output is a dictionary. Identifiers that were not found are stored in the keys ‘failedIds’. Succesful queries are stored in the ‘results’ key that is a list of dictionaries with two keys set to ‘from’ and ‘to’. The ‘from’ key should be in your input list. The ‘to’ key is the result. Here we have the KEGG identifier recognised by its prefix ‘hsa:’, which is for human. Sometimes the output (‘to’) it is more complicated. Consider the following example:
u.mapping("UniParc", "UniProtKB", 'UPI0000000001,UPI0000000002')
You will see that the UniParc results is more complex than just an identifier.
See
valid_mappingattribut for list of valid mapping identifiers.Note that according to Uniprot (June 2022), there are various limits on ID Mapping Job Submission:
Limit
Details
100,000
Total number of ids allowed in comma separated param ids in /idmapping/run api
500,000
Total number of “mapped to” ids allowed
100,000
Total number of “mapped to” ids allowed to be enriched by UniProt data
10,000
Total number of “mapped to” ids allowed with filtering
Changed in version 1.1.1: to return a dictionary instaed of a list
Changed in version 1.1.2: the values for each key is now made of a list instead of strings so as to store more than one values.
Changed in version 1.2.0: input query can also be a list of strings instead of just a string
Changed in version 1.3.1::: use http_post instead of http_get. This is 3 times faster and allows queries with more than 600 entries in one go.
- quick_search(query, include_isoforms=False, sort='score', limit=None)[source]¶
a specialised version of
search()This is equivalent to:
u = uniprot.UniProt() u.search(query, frmt='tsv', sort="score", limit=None)
- Returns
a dictionary.
- retrieve(uniprot_id, frmt='json', database='uniprot', include=False)[source]¶
Search for a uniprot ID in UniProtKB database
- Parameters
- Returns
if the parameter uniprot_id is string, the output will be a a list of identifiers is provided, the output is also a list otherwise, a string. The content of the string of items in the list depends on the value of frmt.
>>> u = UniProt() >>> res = u.retrieve("P09958", frmt="txt") >>> fasta = u.retrieve(['P29317', 'Q5BKX8', 'Q8TCD6'], frmt='fasta') >>> print(fasta[0])
Changed in version 1.10: the xml format is now returned as raw XML. It is not interpreted anymore. The RDF has now an additional option to include data from referenced data sets directly in the returned data (set include=True parameter). Default output format is now set to json.
- search(query, frmt='tsv', columns=None, include_isoforms=False, sort='score', compress=False, limit=None, offset=None, maxTrials=10, database='uniprotkb')[source]¶
Provide some interface to the uniprot search interface.
- Parameters
query (str) – query must be a valid uniprot query. See https://www.uniprot.org/help/query-fields and examples below
frmt (str) – a valid format amongst html, tab, xls, asta, gff, txt, xml, rdf, list, rss. If tab or xls, you can also provide the columns argument. (default is tab)
columns (str) – comma-separated list of values. Works only if fomat is tsv or xls. For UnitProtKB, some possible columns are: id, entry name, length, organism. See also
valid_mappingfor the full list of column keywords.include_isoform (bool) – include isoform sequences when the frmt parameter is fasta. Include description when frmt is rdf.
sort (str) – by score by default. Set to None to bypass this behaviour
compress (bool) – gzip the results
limit (int) – Maximum number of results to retrieve.
offset (int) – Offset of the first result, typically used together with the limit parameter.
maxTrials (int) – this request is unstable, so we may want to try several time.
To obtain the list of uniprot ID returned by the search of zap70 can be retrieved as follows:
>>> u.search('zap70+AND+organism_id:9606') >>> u.search("zap70+AND+taxonomy_id:9606", frmt="tsv", limit=3, ... columns="entry_name,length,id, gene_names") Entry name Length Entry Gene names CBLB_HUMAN 982 Q13191 CBLB RNF56 Nbla00127 CBL_HUMAN 906 P22681 CBL CBL2 RNF55 CD3Z_HUMAN 164 P20963 CD247 CD3Z T3Z TCRZ
other examples:
>>> u.search("ZAP70+AND+organism_id:9606", limit=3, columns="id,xref_pdb")
You can also do a search on several keywords. This is especially useful if you have a list of known entry names.:
>>> u.search("ZAP70_HUMAN+OR+CBL_HUMAN", frmt="tsv", limit=3, ... columns="entry name,length,id, genes") Entry name Length Entry Gene names
Finally, note that when you search for a query, you may have several hits:
>>> u.search("P12345)
including the ID P12345 but also related entries. If you need only the entry that perfectly match the query, use:
>>> u.search("accession:P12345")
This was provided from a user issue that was solved here: https://github.com/cokelaer/bioservices/issues/122
Warning
some columns although valid may not return anything, not even in the header: ‘score’, ‘taxonomy’, ‘tools’. this is a uniprot feature, not bioservices.
Changed in version 1.10: Due to uniprot API changes in June 2022:
parameter ‘include’ is not named ‘include_isoform
default parameter ‘tab’ is now ‘tsv’ but does not change the results
- uniref(query)[source]¶
Calls UniRef service
This is an alias to
retrieve()>>> u = UniProt() >>> u.uniref("Q03063")
Another example from https://github.com/cokelaer/bioservices/issues/121 is the combination of uniprot and uniref filters:
u.uniref("uniprot:(ec:1.1.1.282 taxonomy_name:bacteria reviewed:true)")
Changed in version 1.10: due to uniprot API changes in June 2022, we now return a json instead of a pandas dataframe.
- property valid_mapping¶
8.32. DBFetch¶
Interface to DBFetch web service
What is DBFetch
“DBFetch allows you to retrieve entries from various up-to-date biological databases using entry identifiers or accession numbers. This is equivalent to the CGI based dbfetch service. Like the CGI service a request can return a maximum of 200 entries.”
—From http://www.ebi.ac.uk/Tools/webservices/services/dbfetch , Dec 2012
- class DBFetch(verbose=False)[source]¶
Interface to DBFetch service
>>> from bioservices import DBFetch >>> w = DBFetch() >>> data = w.fetchBatch("uniprot" ,"zap70_human", "xml", "raw")
For more information about the API, check this page: http://www.ebi.ac.uk/Tools/dbfetch/syntax.jsp
Constructor
- Parameters
verbose (bool) – print informative messages
- fetch(query, db='ena_sequence', format='default', style='raw', pageHtml=False)[source]¶
Fetch an entry in a defined format and style.
- Parameters
- Returns
The format of the response depends on the format/style parameter.
from bioservices import DBFetch u = DBFfetch() db.fetch(db="ena_sequence", format="fasta", query="L12344,L12345") db.fetch(db="uniprot", format="fasta", query="P53503")
If db is ommited, the default is ena_sequence. If formatare ommited, the default is EMBL format The default style is raw data.
- get_all_database_info()[source]¶
Get details of all available databases, includes formats and result styles.
- Returns
A list of data structures describing the databases. See
getDatabaseInfo()for a description of the data structure.
- get_database_format_styles(db, format)[source]¶
Get a list of style names available for a given database and format.
- Parameters
- Returns
An array of strings containing the style names.
>>> u.get_database_format_styles("uniprotkb", "fasta") ['default', 'raw', 'html']
- get_database_formats(db)[source]¶
Get list of format names for a given database.
- Parameters
db (str) – valid database name
>>> db.get_database_formats("uniprotkb") ['default', 'annot', 'entrysize', 'fasta', 'gff3', 'seqxml', 'uniprot', 'uniprotrdfxml', 'uniprotxml', 'dasgff', 'gff2']
- get_database_info(db=None)[source]¶
Get details describing specific database (data formats, styles)
- Parameters
db (str) – a valid database.
- Returns
The output can be introspected and contains several attributes
>>> res = u.get_database_info('uniprotkb') >>> print(res['description']) 'The UniProt Knowledgebase (UniProtKB) is the central access point for extensive curated protein information, including function, classification, and cross-references. Search UniProtKB to retrieve everything that is known about a particular sequence.'
- property supported_databases¶
Alias to getSupportedDBs.
8.33. Wikipathway¶
Interface to the WikiPathway service
What is WikiPathway ?
- URL
- REST
- Citation
” WikiPathways is an open, public platform dedicated to the curation of biological pathways by and for the scientific community.”
—From WikiPathway web site. Dec 2012
- class WikiPathways(verbose=True, cache=False)[source]¶
Interface to Pathway service
>>> from bioservices import WikiPathways >>> s = Wikipathway() >>> s.organism # default organism 'Homo sapiens'
Examples:
s.findPathwaysByText('MTOR') s.getPathway('WP1471') s.getPathwaysByOntologyTerm('DOID:344') s.findPathwaysByXref('P45985')
The methods that require a login are not implemented (
login(),updatePathway(),removeCurationTag(),saveCurationTag(),createPathway())Methods not implemented at all:
u’getCurationTagHistory’: No API found in Wikipathway web page
u’getRelations’: No API found in Wikipathway web page
Constructor
- Parameters
verbose (bool) –
- createPathway(gpmlCode, authInfo)[source]¶
Create a new pathway on the WikiPathways website with a given GPML code.
Warning
Interface not exposed in bioservices.
Note
To create/modify pathways via the web service, you need to have an account with web service write permissions. Please contact us to request write access for the web service.
- Parameters
gpml (str) – The GPML code.
auth (object WSAuth) – The authentication info.
- Returns
WSPathwayInfo The pathway info for the created pathway (containing identifier, revision, etc.).
- findInteractions(query)[source]¶
Find interactions defined in WikiPathways pathways.
- Parameters
query (str) – The name of an entity to find interactions for (e.g. ‘P53’)
- Returns
list of dictionaries
res = w.findInteractions("P53")
- findPathwaysByLiterature(query)[source]¶
Find pathways by their literature references.
- Parameters
query (str) – The query, can be a pubmed id, author name or title keyword.
- Returns
dictionary with Pathway as keys
res = s.findPathwaysByLiterature(18651794)
- findPathwaysByText(query, species=None)[source]¶
Find pathways using a textual search on the description and text labels of the pathway objects.
The query syntax offers several options:
Combine terms with AND and OR. Combining terms with a space is equal to using OR (‘p53 OR apoptosis’ gives the same result as ‘p53 apoptosis’).
Group terms with parentheses, e.g. ‘(apoptosis OR mapk) AND p53’
You can use wildcards * and ?. * searches for one or more characters, ? searches for only one character.
Use quotes to escape special characters. E.g. ‘“apoptosis*”’ will include the * in the search and not use it as wildcard.
This function supports REST-style invocation. Example: http://www.wikipathways.org/wpi/webservice/webservice.php/findPathwaysByText?query=apoptosis
- Parameters
- Returns
Array of WSSearchResult An array of search results.
s.findPathwaysByText(query="p53 OR mapk",species='Homo sapiens')
Warning
AND or OR must be in big caps
- findPathwaysByXref(ids, codes=None)[source]¶
Find pathways by searching on the external references of DataNodes.
- Parameters
ids (str string) – One or mode DataNode identifier(s) (e.g. ‘P45985’). Datanodes can be (gene/protein/metabolite identifiers). For one node, you can use a string (or number) or list of one identifier. you can also provide a list of identifiers.
codes (str) – You can restrict the search to a specific database. See http://developers.pathvisio.org/wiki/DatabasesMapps#Supporteddatabasesystems for details. Examples are “L” for entrez gene, “En” for ensembl. See also the note here below for multiple identifiers/codes.
- Returns
a dictionary
>>> s.findPathwaysByXref(ids="P45985") >>> s.findPathwaysByXref(ids="P45985", codes="L") >>> s.findPathwaysByXref(ids=["P45985"], codes=["L"]) >>> s.findPathwaysByXref(ids=["P45985", "ENSG00000130164"], codes=["L", "En"])
Note that in the last example, we specify multiple ids and codes parameters to query for multiple xrefs at once. In that case, the number of ids and codes parameters should match. Moreover, they will be paired to form xrefs, so P45985 is searched for in the “L” database while “ENSG00000130164” is searched for in the En” database only.
- getColoredPathway(pathwayId, filetype='svg', revision=0, color=None, graphId=None)[source]¶
Get a colored image version of the pathway.
- Parameters
- Returns
Binary form of the image.
Todo
graphId, color parameters
- getCurationTags(pathwayId)[source]¶
Get all curation tags for the given pathway.
- Parameters
pathwayId (str) – the pathway identifier.
- Returns
Array of WSCurationTag. The curation tags.
s.getCurationTags("WP4")
- getCurationTagsByName(name)[source]¶
Get all curation tags for the given tag name.
Use this method if you want to find all pathways that are tagged with a specific curation tag.
- Parameters
tagName (str) – The tag name.
- Returns
Array of WSCurationTag. The curation tags (one instance for each pathway that has been tagged).
s.getCurationTagsByName("Curation:FeaturedPathway")
- getOntologyTermsByPathway(pathwayId)[source]¶
Get a list of ontology terms for a given pathway.
- Parameters
pathwayId (str) – the pathway identifier.
- Returns
Array of WSOntologyTerm. The ontology terms.
s.getOntologyTermsByPathway("WP4")
- getPathway(pathwayId, revision=0)[source]¶
Download a pathway from WikiPathways.
- Parameters
- Returns
The pathway as a dictionary. The pathway is stored in gpml format.
s.getPathway("WP2320")
- getPathwayAs(pathwayId, filetype='png', revision=0)[source]¶
Download a pathway in the specified file format.
- Parameters
- Returns
The file contents
Changed in version 1.3.0: return raw output of the service without any parsing
Note
use
savePathwayAs()to save into a file.
- getPathwayHistory(pathwayId, date)[source]¶
Get the revision history of a pathway.
- Parameters
- Returns
The revision history.
Warning
seems unstable does not return the results systematically.
s.getPathwayHistory("WP4", 20110101000000)
- getPathwayInfo(pathwayId)[source]¶
Get some general info about the pathway.
- Parameters
pathwayId (str) – the pathway identifier.
- Returns
The pathway info.
>>> from bioservices import * >>> s = Wikipathway() >>> s.getPathwayInfo("WP2320")
- getPathwaysByOntologyTerm(terms)[source]¶
Get a list of pathways tagged with a given ontology term.
- Parameters
terms (str) – the ontology term identifier.
- Returns
dataframe with pathways infomation.
>>> from bioservices import WikiPathways >>> s = Wikipathway() >>> s.getPathwaysByOntologyTerm('PW:0000724')
- getPathwaysByParentOntologyTerm(term)[source]¶
Get a list of pathways tagged with any ontology term that is the child of the given Ontology term.
- Parameters
term (str) – the ontology term identifier.
- Returns
List of WSPathwayInfo The pathway information.
- getRecentChanges(timestamp)[source]¶
Get the recently changed pathways.
- Parameters
timestamp (str) – Only get changes from after this time. Timestamp format: yyyymmddMMHHSS (string or number)
- Returns
The changed pathways in XML format
s.getRecentChanges(20110101000000)
Todo
interpret XML
- listPathways(organism=None)[source]¶
Get a list of all available pathways.
- Parameters
organism (str) – If provided, the data is filtered to keep only the organism provided, which must be a valid name (check out
organismattribute)- Returns
dataframe. Index are the pathways identifiers (e.g. WP1)
(Source code, png, hires.png, pdf)
- login(usrname, password)[source]¶
Start a logged in session using an existing WikiPathways account.
Warning
Interface not exposed in bioservices.
This function will return an authentication code that can be used to excecute methods that need authentication (e.g. updatePathway).
- property organism¶
Read/write attribute for the organism
- organisms¶
Get a list of all available organisms.
- removeCurationTag(pathwayId, name)[source]¶
Remove a curation tag from a pathway.
Warning
Interface not exposed in bioservices.
- saveCurationTag(pathwayId, name, revision)[source]¶
Apply a curation tag to a pathway. This operation will overwrite any existing tag with the same name.
Warning
Interface not exposed in bioservices.
- Parameters
pathwayId (str) – the pathway identifier.
- savePathwayAs(pathwayId, filename, revision=0, display=True)[source]¶
Save a pathway.
- Parameters
pathwayId (str) – the pathway identifier.
filename (str) – the name of the file. If a filename extension is not provided the pathway will be saved as a pdf (default).
revisionNumb (int) – the revision number of the pathway (use ‘0 for most recent version).
display (bool) – if True the pathway will be displayed in your browser.
Note
Method from bioservices. Not a WikiPathways function
Changed in version 1.7: return PNG by default instead of PDF. PDF not working as of 20 Feb 2020 even on wikipathway website.
- showPathwayInBrowser(pathwayId)[source]¶
Show a given Pathway into your favorite browser.
- Parameters
pathwayId (str) – the pathway identifier.
- updatePathway(pathwayId, describeChanges, gpmlCode, revision=0)[source]¶
Update a pathway on WikiPathways website with a given GPML code.
Warning
Interface not exposed in bioservices.
Note
To create/modify pathways via the web service, you need to have an account with web service write permissions. Please contact us to request write access for the web service.
- Parameters
pwId (str) – The pathway identifier.
description (str) – A description of the modifications.
gpml (str) – The updated GPML code.
revision (int) – The revision number of the version this GPML code was based on. This is used to prevent edit conflicts in case another client edited the pathway after this client downloaded it.
WSAuth_auth (object) – The authentication info.
- Returns
Boolean. True if the pathway was updated successfully.
9. Applications and extra tools¶
Web services have lots of overlap amongst themselves. For instance, fetching a FASTA sequence can be done using many different services. Yet, once a FASTA is retrieved, one may want to perform additional tasks or save the FASTA into a file or whatever repetitive functionalities not included in Web Services anymore.
The goal of this sub-package is to provide convenient tools, which are not web services per se but that makes use of one or several Web Services already available within BioServices.
Warning
this is experimental and was added in version 1.2.0 so it may change quite a lot.
9.1. Peptides¶
9.2. FASTA¶
- class FASTA[source]¶
Dedicated class to manipulates FASTA sequence(s)
Here is a FASTA file example:
>sp|P43408|KADA_METIG Adenylate kinase OS=Methanotorris igneus GN=adkA PE=1 SV=2 MKNKVVVVTGVPGVGGTTLTQKTIEKLKEEGIEYKMVNFGTVMFEVAKEEGLVEDRDQMR KLDPDTQKRIQKLAGRKIAEMAKESNVIVDTHSTVKTPKGYLAGLPIWVLEELNPDIIVI VETSSDEILMRRLGDATRNRDIELTSDIDEHQFMNRCAAMAYGVLTGATVKIIKNRDGLL DKAVEELISVLK
The format is made of a header and a sequence. Any FASTA can be read and the pair of header/sequence retrieved from the
sequenceandheaderattributes. However, headers differ from one database to another one and interpretation is not implemented except for SWISS-PROT. Identifiers can be retrieved whatsoever.You can read a FASTA sequence from a local file or download one from UniProt
>>> from bioservices.apps.fasta import FASTA >>> f = FASTA() >>> f.load("P43403") >>> acc = f.accession # the accession (P43403) >>> fasta = f.fasta # raw FASTA string >>> seq = f.sequence # the sequence itself >>> header = f.header # the header itself >>> identifier = f.identifier
You can also get a dataframe also using Pandas library.:
>>> f.df
The columns stored in the dataframe encompase:
Accession that is taken from the header (e.g., P43403 from uniprot)
Sequence, a copy of the FASTA sequence
Size, the length of the sequence.
Database, the database type found in the header (e.g., sp for SWISS-PROT; see below for a list of database and their header format).
Some column such as Organism are filled only for some database
Identififers is the begining of the header.
See also
MultiFASTAfor multi FASTA manipulation.List of identifiers corresponding to different databases.
GenBank
gi|gi-number|gb|accession|locus
EMBL Data Library
gi|gi-number|emb|accession|locus
DDBJ, DNA Database of Japan
gi|gi-number|dbj|accession|locus
NBRF PIR
pir||entry
Protein Research Foundation
prf||name
SWISS-PROT
sp|accession|name
Brookhaven Protein Data Bank (1)
pdb|entry|chain
Brookhaven Protein Data Bank (2)
entry:chain|PDBID|CHAIN|SEQUENCE
Patents
pat|country|number
GenInfo Backbone Id
bbs|number
General database identifier
gnl|database|identifier
NCBI Reference Sequence
ref|accession|locus
Local Sequence identifier
lcl|identifier
The :meth::load_fasta relies on UniProt service.
- property PE¶
returns PE keyword found in the header if any
- property SV¶
returns SV keyword found in the header if any
- property accession¶
- property dbtype¶
- property df¶
- property entry¶
returns entry only
- property fasta¶
returns FASTA content
- property gene_name¶
returns gene name from GN keyword found in the header if any
- get_fasta(id_)[source]¶
Fetches FASTA from uniprot and loads into attrbiute
fasta- Parameters
id (str) – a given uniprot identifier
- Returns
the FASTA contents
- property header¶
returns header only
- property identifier¶
- known_dbtypes = ['sp', 'gi']¶
- load_fasta(id_)[source]¶
Fetches FASTA from uniprot and loads into attribute
fasta- Parameters
id (str) – a given uniprot identifier
- Returns
nothing
Note
same as
get_fasta()but returns nothing
- property name¶
- property organism¶
returns organism from OS keyword found in the header if any
- read_fasta(filename)[source]¶
Reads a FASTA file and loads it
Type:
>>> f = FASTA() >>> f.read_fasta(filename) >>> f.fasta
- Returns
nothing
Warning
If more than one FASTA is contained in the file, an error is raised
- property sequence¶
returns the sequence only
- class MultiFASTA[source]¶
Class to manipulate several several FASTA items
Here, we load some FASTA using UniProt web service:
>>> from bioservices import MultiFASTA >>> mf = MultiFASTA() >>> mf.load_fasta("P43408") >>> mf.load_fasta("P21318")
You can then get back to your accession entries as follows
>>> mf.ids ['P43408', 'P21318']
And the sequences in the same order can be accessed:
>>> len(mf) 2
Each FASTA is stored in
fasta, which is a dictionary where each values is an instance ofFASTA:>>> print(mf._fasta["P43408"].fasta) >sp|P43408|KADA_METIG Adenylate kinase OS=Methanotorris igneus GN=adkA PE=1 SV=2 MKNKVVVVTGVPGVGGTTLTQKTIEKLKEEGIEYKMVNFGTVMFEVAKEEGLVEDRDQMR KLDPDTQKRIQKLAGRKIAEMAKESNVIVDTHSTVKTPKGYLAGLPIWVLEELNPDIIVI VETSSDEILMRRLGDATRNRDIELTSDIDEHQFMNRCAAMAYGVLTGATVKIIKNRDGLL DKAVEELISVLK
The most convenient way to access to all data is to use the dataframe attribute:
>>> mf.df.Sequence
>>> from bioservices.apps import MultiFASTA >>> f = MultiFASTA() >>> f.load_fasta(["P43403", "P43410"]) >>> f.df.Size.hist()
(Source code, png, hires.png, pdf)
- property df¶
- property fasta¶
Returns all FASTA instances
- hist_size(**kwds)¶
- property ids¶
returns list of keys/accession identifiers
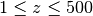 )
)